#molecular phylogeny
Text
Pandas, Kitzmiller, and the frozen frog fallacy
By Paul Braterman
January 4, 2023 13:00 MST
Paul Braterman is Professor Emeritus at the University of North Texas and Honorary Senior Research Fellow in Chemistry at the University of Glasgow
This Kitzmas was different. For the first time, the Discovery Institute allowed the anniversary of Kitzmiller v. Dover Area School District to pass without complaining about the verdict. Perhaps they are…

View On WordPress
#Creationism#Cytochrome C#Discovery Institute#Frozen Frog Fallacy#Intelligent design#Kitzmiller v Dover#Molecular phylogeny#Of Pandas and People
0 notes
Text
Scientists Discover How New Caledonia Birthed Over 40 Unique Palm Species
The unique palm flora of New Caledonia contains over 40 species found nowhere else on Earth. However, the complex geological history of the island and lack of detailed phylogenetic studies have left major questions around how this diversity arose. A new multi-year study led by Victor Pérez-Calle recently published in Annals of Botany has constructed the most comprehensive phylogenetic tree of New…

View On WordPress
#Arecaceae#Areceae#biogeography#molecular dating#New Caledonia#phylogeny#speciation#target sequence capture#ultramafic
0 notes
Text
Alex Wild got me interested in attaphila fungicola with this first super cute photo of the myrmecophilous roaches that live with leaf cutter (fungi farming) ants.

I think their habit of harmlessly hitching rides on ants could inspire a line of adorable felt pillboxes. Can you see it?


#pillbox hat#ants#antposting#insects#bugblr#ant#myrmecology#roaches#invertebrates#fashion#inspiration#antblr#classic fashion#pillbox#haberdashery#hats#bugs#myrmecophilous roaches#leaf cutter ants#leafcutter ants#atta
283 notes
·
View notes
Photo
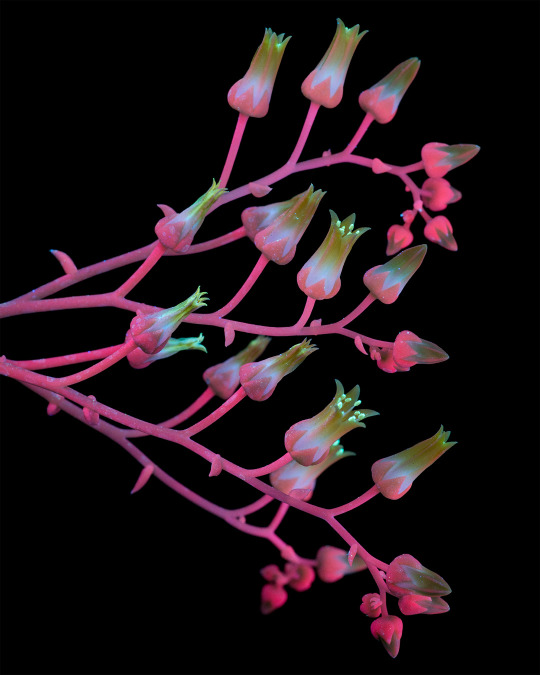
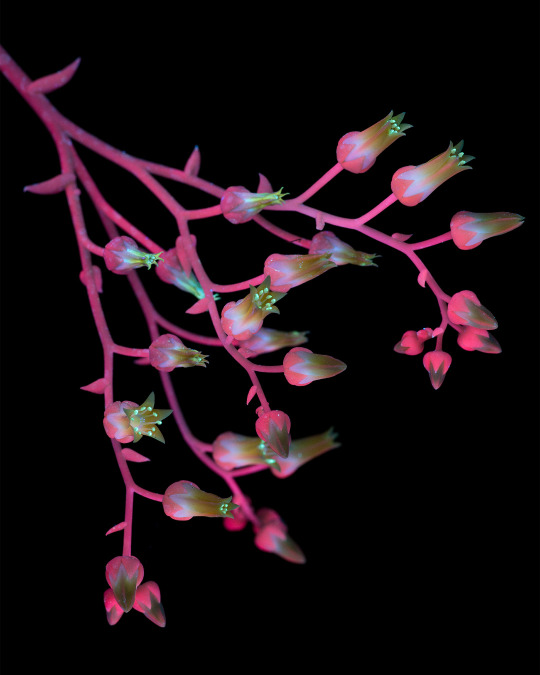





Dudleya cymosa ssp. pumila is a succulent native and endemic to California, meaning it cannot be found growing naturally anywhere else in the world. This plant is most often found on north-facing canyon walls and roadcuts throughout the transverse ranges (a series of mountains that run east-west through CA instead of the usual north-south.) Dudleya in general have special adaptations which allow them to survive challenging environments, in this case growing in the wet but coldest part of the year and surviving a long, very hot, and very dry summer.
Even in this subspecies there are many included forms, with varying traits such as flower color, leaf farina, altitude, etc., which goes in hand with Dudleya being a genus of complicated and still-developing taxonomy with quite a lot of undescribed species. Molecular phylogeny is making this possible by sampling genetic material to determine evolutionary relationships. This process is further complicated by the fact that Dudleya is a highly poached genus and many populations of unique plants are threatened by sprawling human development and introduced herbivores before they even have a chance to be recognized.
Dudleya is one of my favorite genera of plants, so expect to see more of them in my posts in the future!
296 notes
·
View notes
Note
re: your last post about career options in entomology, consider this an invitation to talk about what you think about molecular systematics and its role in taxonomy/phylogeny. i'm an undergrad currently in a lab focused on the phylogeny of benthic marine inverts, and the vast bulk of our work is based on molecular data. i'm interested in hearing your thoughts on the topic!
i should be clear and say that i have no inherent problems with molecular systematics as an approach to taxonomy, and in many cases it can be a tremendous boon to the field and pretty much the only way to untangle some particularly gnarly taxonomic knots.
i guess my problem arises from how the ascendancy of molecular systematics is crowding out traditional morphological taxonomy, to the point where, like i was complaining in my last post, many subdisciplines are disappearing because nobody is replacing the aging and dying experts on many groups of insects and other organisms. i think that very few people who do molecular systematics would themselves argue that they should replace traditional taxonomists, but i also think that academic administrators and funding agency bureaucrats see the difference in "efficiency" between one scientist who needs years if not decades to publish a comprehensive monograph on their study group, and another who can slam a bunch of samples through a machine and describe a hundred new species in a semester, and would prefer to fund the latter. i'm being unfairly reductive here of course, but my point is that in the publish-or-perish culture of academia, being able to churn out papers faster will always be rewarded and tilts the game way towards molecular taxonomists. even incoming grad students who want to study to become morphological taxonomists are having a harder time securing the funds to do so.
i just worry about what things are going to look like in even 10 or 20 years from now when the last of the old-timers have died off and very few have been replaced. even if you can technically identify an organism of interest through sequencing it's CO1 gene or whatever, that's no replacement for having an actual expert who can way more quickly and authoritatively tell you what it is without having to be connected to Genbank.
(far more minor, but i also think that molecular work places far too great of an emphasis on dogmatic phylogenetic taxonomizing over more practical groupings. this is pure old man grumbling but i absolutely can't stand how every few years someone recategorizes some group of insects based on their genetic sequences and suddenly i gotta relearn a bunch of new taxonomies and relabel a bunch of shit because every group's just gotta be 100% monophyletic. fuck monophyly! i will not elaborate on this!)
anyway back to my main point, again i'm sure i'm being reductive and unfair and there are absolutely labs that make use of both morphological and molecular taxonomic methods, but it's just a worrying trend to me. i just can't help but feel like the ascendancy of molecular systematics is of a piece with the inexorable automation of so many other jobs and disciplines, like a robot replacing the craftsman it was ostensibly designed to assist.
#entomology#nobody is allowed to yell at me about my opinions on taxonomy#im just a little guy#and im very sensitive
52 notes
·
View notes
Note
https://vxtwitter.com/albertonykus/status/1757175498378219573?s=20
Curious as to your thoughts on the legitimacy of this study? Cause it doesn't seem to line up with what we know of previous studies and the fossil record
I study how birds adapted to the PETM and I am extremely skeptical about their claim that this water bird clade they recovered emerged right after - one of the best bird fossil records from the Paleocene we have is stem-penguins. So clearly, they were around before then. And they don't even acknowledge that in the paper!
The phylogeny is interesting but as always I am a major skeptic about molecular clocks and this is no exception, given, again, the fossils that directly contradict the damn conclusion.
Other than that I haven't had time to go in depth enough, but those are my initial thoughts
Side note, when will a stable Neoavian phylogeny return from the war
The paper, for those curious:
12 notes
·
View notes
Text
2023 Reading Log pt. 14
Where the hell did November go?

66. New World Monkeys: The Evolutionary Odyssey by Alfred L. Rosenberger. In the introduction, the author laments that there aren’t any good books outlining the evolution and ecology of the New World Monkeys. If that's the case, there still aren’t. This book does alright by the ecology part—it has good summaries of the anatomy, behavior and feeding interactions of the covered monkeys. But the evolution is a mess. Rosenberger’s take on the evolutionary relationships between the animals covered here is iconoclastic, to say the least. He distrusts molecular phylogeny, uses synapomorphic characters that are basically just vibes, and has an entire chapter dedicated to lambasting the idea that any mammals could disperse across the Atlantic Ocean from Africa to South America (the consensus explanation) in favor of a hypothesized trek through Greenland and North America that has no evidence and still requires open ocean crossings. This was an incredibly frustrating experience to read, because there’s enough good content among the dross that I didn’t want to just abandon it.

67. Seaweeds of the World by John H. Bothwell. The weakest of Princeton University Press’ “X of the World” series. For one thing, the subtitle is usually “A Guide to Every Family”, whereas here it’s “A Guide to Every Order”. The book’s general coverage of seaweeds is pretty good—it explains why “seaweed” is a polyphyletic category but still useful in common English, explains the anatomy and the complex life histories of seaweeds. But the actual coverage of groups is lacking. Again, it doesn’t cover every family. And it’s more interested in seaweeds of economic importance than it is in their actual ecologies. Plus the writing is just kinda boring. This is the first entry in this series I do not recommend.

68. Lapidarium: The Secret Lives of Stones by Hettie Judah. Now this is more like it! This book is a series of short essays about stones and their cultural impact. I’m a sucker for cultural histories in general, and this is a very good one. I especially liked that it doesn’t just cover gemstones, as I originally expected, but also stones used in art and architecture, resources like coal, and the use of earthworks in religion. The focus is much more on the culture than the geology, but the book does discuss things like deposition of sediments and how metamorphic rocks yield gemstones in explaining why certain places have certain rocks. The book is also lovely to look at, with minimalist bands of color along the sides of the pages in the hues of the stones covered in that chapter.

69. Monsters and Monarchs: Serial Killers in Classical Myth and History by Debbie Felton. I was excited for this one. I had read Felton’s chapter in Monster Anthropology, which suggested that Greek traveler’s tales about werewolves and the murderous robbers encountered by Theseus in myth were both expressions of cultural fears about serial killers. Unfortunately, that article already covered the bulk of Felton’s actual argument and evidence, and this book is those 20 pages fluffed to 200. The only other really good material is some coverage of the distinction between Greek and Roman attitudes towards law and order, and what “counted” as murder in each society. The rest of it is handwaving and extrapolation from very little data, with just about every instance of mass killing that we have records of, from political uprisings to court intrigue, being taken as the work of a possible serial killer. Plus, the author is a Freudian, so we have to hear about coded references to rape and sexual violence in stories where there really aren’t any. Sometimes a bed where you get your legs cut off is just a bed.

70. Cult of the Dead: A Brief History of Christianity by Kyle Smith. They say you can’t judge a book by its cover, of course, but that title and that cover made an instant sale for me. I’m glad it did, too, because this is a good one. An explanation of the importance of martyrdom to Christianity, it does an excellent job of explaining why, exactly, so many people were willing and eager to die for their faith, and how this persisted in building a persecution complex among the dominant European religion for centuries. The book avers from discussing the present day for the most part, tapering off with the work of reformist Catholics poring over the many, many legends about saints and trying to determine which, if any of them, represent actual historical events rather than religious fictions. Other topics covered include the trade in relics, the role in martyrologies in shaping the modern calendar, and how women could most easily play a role in the Church through the mortification of the flesh. The book is eminently readable and very well illustrated.
#reading log#christianity#comparative religion#seaweed#classical mythology#classical greece#classical rome#new world monkeys#evolution#ecology#geology#cultural history
8 notes
·
View notes
Text
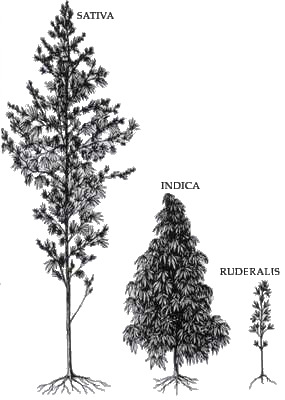
Contrary to what every stoner will excitedly tell you, and what you will see in most every dispensary, there is technically only one species of Cannabis, Cannabis ruderalis.
"sativa" means cultivated, as in a cultivar of ruderalis
rūdus, rūder- (“rubble, debris”) + -ālis "to grow" --- Interestingly implying it grows in disturbed areas (like a "weed").
"indica", modern Latin for “of India."
Taxonomy has changed a lot since they were initially identified, and the advent of molecular phylogeny
Phylogenetic studies showed both Cannabis "sativa" and "indica" share the common ancestor Cannabis ruderalis and that "indica" is a cultivar of "sativa" which is a cultivated variety of Cannabis ruderalis
For Cannabis sativa to be its own species it would need at least two common ancestors, Cannabis ruderalis being the only known en-situ unaltered native species of Cannabis
Unless we discover another
#sorry stoners#botany#taxonomy#i actually love cannabis#i have worked with cannabis for over 4 years
60 notes
·
View notes
Text
Curated List of My Tumblr Posts on Paleontology, Classification, Doraemon, etc.
I don’t currently take asks on this blog, but over the years I’ve accumulated a good amount of material on here that I essentially haven’t posted anywhere else. Given that finding old posts on Tumblr can be a difficult and arduous task, I’ve decided to assemble a curated selection of links to my posts that I think are of particular interest (whether on a general or personal level). These are often posts that have been especially popular, posts that I especially enjoyed writing, or both. Yes, I’m including the Doraemon reviews. 😛 For ease of access, I have also replicated this list on a page on this blog, which I will try to keep updated as I deem necessary.
Paleontology Resources
What are some good introductory resources for dinosaur paleontology?
What are some good YouTube channels about paleontology? (needs updating)
Some ways to access paywalled scientific papers (and why I won’t repost published phylogenetic figures on @new-dinosaurs)
What should one major in college to get into paleontology?
How do I make phylogenetic diagrams?
How to subscribe to the Dinosaur Mailing List
Specific Dinosaur Questions
How flexible are dinosaur tails?
What types of dinosaurs have feathers?
Wasn’t there a study showing that feathers were not an ancestral trait of dinosaurs?
How reliable are melanosomes for reconstructing the colors of extinct dinosaurs?
Why does paleoart of feathered dinosaurs tend to show the tip of snout unfeathered?
Is it true that non-avian dinosaurs couldn’t roar?
Which dinosaurian herbivores are foregut fermenters and which ones are hindgut fermenters?
When did dinosaurs evolve hollow bones?
Could sauropods swim?
Can any theropods pronate their hands? (And how about other reptiles?)
Were giant maniraptoriforms likely to have been featherless?
Did flightless non-avialan pennaraptorans have feather barbules?
Would Microraptor and Anchiornis have had trouble walking due to their large hindlimb feathers?
What do we know about the social and reproductive behaviors of dromaeosaurids?
Did bird ancestors evolve flight from the ground up or trees down? How might flight have evolved from the ground up? Do we have extant analogues for such a process?
How do we know birds are actually dinosaurs, and that we haven’t been misled by convergent evolution?
Why do birds have backward-pointing dewclaws?
What is the most likely phylogeny of modern birds?
Are eider ducks the fastest animals in the world?
Were there penguins in the Cretaceous?
Why do bateleur eagles have short tail feathers?
Do all owls have asymmetrical ears?
Are falcons closely related to parrots?
How many times did poison evolve in songbirds?
Taxonomy, Nomenclature, and Phylogenetics
How do you pluralize genus/species names?
What is the phylogenetic species concept?
What is wrong with ranked/Linnaean taxonomy?
If birds are reptiles, shouldn’t tetrapods be considered fish?
Should we avoid calling birds dinosaurs because they were not traditionally called dinosaurs?
How is phylogenetic nomenclature reconciled with the fact that species must have evolved from other species?
What is a synapomorphy and how do we identify one?
How often do morphological and molecular phylogenetics agree?
General/Other Biology
Should the study of birds be included under herpetology?
Why is monogamy more common in birds than in mammals?
We don’t know what ichthyosaurs and sauropterygians are (needs updating)
How do reptiles drink?
How to identify a rodent skull
Why do mammals have ear flaps?
Could a mammal evolve as many neck vertebrae as a bird?
How can you tell the position of an animal’s ears by looking at its skull?
What is the difference between mesothermy and endothermy?
Why has the "Handicap Principle” been disputed?
Does any organic material remain in fossils?
Does it worry me that paleontologists will run out of fossils to discover?
What do I think about using humor in scientific outreach?
Just for Fun
Meme about fossil bird books
Fusion is a cheap tactic to make weak reptiles stronger
Meme about horses in geology
What are some works of paleo-fiction that I enjoy? (needs updating)
What are some webcomics about extinct animals that I enjoy? (needs updating)
List of science-themed music artists (needs updating)
Phylogeny (“Under the Sea” parody)
We don’t talk about Spino
Are there non-talking horses in Equestria (from My Little Pony: Friendship is Magic)?
Hilda and the nature of revelations
Doraemon
Movie review: Nobita’s Dinosaur (1980) and Nobita’s Dinosaur (2006)
Movie review: The Records of Nobita, Spaceblazer (1981) and The New Record of Nobita’s Spaceblazer (2009)
Movie review: Nobita and the Haunts of Evil (1982) and New Nobita’s Great Demon (2014)
Movie review: Nobita and the Castle of the Undersea Devil (1983)
Movie review: Nobita’s Great Adventure into the Underworld (1984) and Nobita’s New Great Adventure into the Underworld (2007)
Movie review: Nobita’s Little Star Wars (1985) and Nobita’s Little Star Wars 2021 (2022)
Movie review: Nobita and the Steel Troops (1986) and Nobita and the New Steel Troops (2011)
Movie review: Nobita and the Knights on Dinosaurs (1987)
Movie review: The Record of Nobita’s Parallel Visit to the West (1988)
Movie review: Nobita and the Birth of Japan (1989) and Nobita and the Birth of Japan (2016)
Movie review: Nobita and the Animal Planet (1990)
Movie review: Nobita’s Dorabian Nights (1991)
Movie review: Nobita and the Kingdom of Clouds (1992)
Movie review: Nobita and the Tin Labyrinth (1993)
Movie review: Nobita’s Three Visionary Swordsmen (1994)
Movie review: Nobita’s Diary on the Creation of the World (1995)
Movie review: Nobita and the Galaxy Super-express (1996)
Movie review: Nobita and the Spiral City (1997)
Movie review: Nobita’s Great Adventure in the South Seas (1998)
Movie review: Nobita Drifts in the Universe (1999)
Movie review: Nobita and the Legend of the Sun King (2000)
Movie review: Nobita and the Winged Braves (2001)
Movie review: Nobita in the Robot Kingdom (2002)
Movie review: Nobita and the Windmasters (2003)
Movie review: Nobita in the Wan-Nyan Spacetime Odyssey (2004)
Movie review: Nobita and the Green Giant Legend (2008)
Movie review: Nobita’s Great Battle of the Mermaid King (2010)
Movie review: Nobita and the Island of Miracles (2012)
Movie review: Nobita’s Secret Gadget Museum (2013)
Movie review: Stand by Me Doraemon (2014)
Movie review: Nobita’s Space Heroes (2015)
Movie review: Nobita’s Great Adventure in the Antarctic Kachi Kochi (2017)
Movie review: Nobita’s Treasure Island (2018)
Movie review: Nobita’s Chronicle of the Moon Exploration (2019)
Movie review: Nobita’s New Dinosaur (2020)
Movie review: Stand by Me Doraemon 2 (2020)
Ranking the Doraemon movies (1980–2022)
Where to find Doraemon in English
20 notes
·
View notes
Photo

A consolidated phylogeny of snail-eating snakes (Serpentes, Dipsadini), with the description of five new species from Colombia, Ecuador, and Panama
Alejandro Arteaga, Abel Batista
Abstract
A molecular phylogeny of the Neotropical snail-eating snakes (tribe Dipsadini Bonaparte, 1838) is presented that includes 60 of the 133 species currently recognized.
There is morphological and phylogenetic support for four new species of Sibon Fitzinger, 1826 and one of Dipsas Laurenti, 1768, which are described here based on their unique combination of molecular, meristic, and color pattern characteristics.
Plesiodipsas Harvey et al., 2008 is designated as a junior synonym of Dipsas and additional evidence is presented to support the transfer of the genus Geophis Wagler, 1830 to the tribe Dipsadini.
Two of the subspecies of S. nebulatus (Linnaeus, 1758) are elevated to full species status. Insight into additional undescribed cryptic diversity within the S. nebulatus species complex is provided.
Evidence that supports the existence of an undescribed species previously confused with D. temporalis is provided, as well as the first country record of S. ayerbeorum Vera-Pérez, 2019 in Ecuador with a comment on the ontogenetic variation of the latter.
Finally, photographs of Colombian, Ecuadorian, and Panamanian snail-eating snakes are provided.
Read the paper here:
https://zookeys.pensoft.net/article/93601/
#herpetology#science#snake#snail eater#snail eating snake#reptile#animals#nature#south america#central america
17 notes
·
View notes
Text
From Longreads (@longreads):
"The Last of the Fungus
Zhengyang Wang | Nautilus | August 30, 2023 | 4,497 words
Molecular phylogenies and caterpillar fungus are not topics I expected to find riveting until I read Zhengyang Wang’s essay. His story bounces along like a thriller: Mountaintop expeditions, dodgy deals, and even death are part of this fungal world. However, the most gripping thread is Wang’s PhD. Yes, that’s right: He makes his PhD research project on parasitic fungus sound fascinating. The parasite in question is reminiscent of Alien, invading ghost moth caterpillars and taking over their brains until stroma blasts out of their heads and sticks up from the soil. (Wang describes this much more eerily and beautifully.) In China, this stroma is celebrated for helping with a different kind of protrusion and is known as “Himalayan Viagra.” The attributed medical and aphrodisiacal powers (by no means proven) mean the sale of this fungus equates to a massive tenth of Tibet’s gross domestic product. Inevitably, people are attempting industrial farming, and mountain vistas are being devastated as caterpillars are collected to sell to fungus breeders. But it isn’t working. Spraying caterpillars with spores of the parasite O. sinensis does not infect them. Wang’s PhD explains why these centers are failing: The complicated, intricate ecosystems where these hosts and parasites evolve together are impossible to replicate. His research proves the decimation of delicate montane habitats is pointless, but not enough people are reading it. You can. —CW"
2 notes
·
View notes
Text
It's time for one of Sarnida's taxonomy hot takes! There are a bunch of animals that are functionally identical and scientists are like wehhh that's a different animal because of phylogeny and karyotypes 🤓👶 and that's like, ok in a strictly technical context, but then people bring that wretched energy into casual conversation and they should be killed. You know what the difference is between crocodiles and alligators? Basically fucking nothing honestly like if you followed one around for a week you wouldn't tell the difference. "Oh but they're descended from different evolutionary lines we can tell because of the protein sequences" mate the cool video I saw was not on a molecular scale
2 notes
·
View notes
Text
The temperate South American lizard genus Liolaemus is the one of the most widely distributed and species-rich genera of lizards on earth. The genus is divided into two subgenera, Liolaemus sensu stricto (the ‘Chilean group’) and Eulaemus (the ‘Argentino group’), a division that is supported by recent molecular and morphological data. Owing to a lack of reliable fossil data, previous studies have been forced to use either global molecular clocks, a standardized mutation rate adopted from previous studies, or the use of geological events as calibration points. However, simulations indicate that these types of assumptions may result in less accurate estimates of divergence times when clock-like models or mutation rates are violated. We used a multilocus data set combined with a newly described fossil to provide the first calibrated phylogeny for the crown groups of the clade Eulaemus, and derive new fossil-calibrated substitution rates (with error) of both nuclear and mtDNA gene regions for Eulaemus specifically. Divergence date estimates for each of the crown groups and appropriate rate estimates will provide the foundation for understanding rates of speciation, historical biogeography, and phylogeographical history for various clades in one of the most diverse lizard genera in the poorly studied Patagonian region.
5 notes
·
View notes
Text
So last week shared a reflection on a science paper with you guys and I thought it went pretty well. So tonight, let’s talk photosynthesis.
I’m mostly going to be discussing this 2018 paper, “Dating phototrophic microbial lineages with reticulate gene histories.” However, since I’ve also taken classes that discussed this issue at length, I’ll be pulling in some outside knowledge too.
Most people that took high school bio probably remember that chloroplasts, the organelles responsible for photosynthesis in plant cells, have two photosystems: photosystem II (discovered second, but first in sequence) and photosystem I. What you may or may not know is that these photosystems have a really convoluted evolutionary history.
“But wait!” you (hypothetically) say. “These two photosystems are so similar! Isn’t gene duplication the simplest, most parsimonious explanation?” Nope. Probably not
Gene duplication seems like it would be the obvious candidate for How Chloroplasts Ended Up With Two Photosystems, but alas, there’s a pretty big mountain of phylogenetic evidence that this is not the case. There are at least five groups of photosynthetic bacteria that have a homolog for photosystem II OR photosystem I, but not both. None of these are oxygen producing. In other words, both photosystems are distinctive and neither seems to have emerged based on the other. This throws a wrench in things.
Stem-lineage cyanobacteria are considered to be the MRCA (most recent common ancestor) of chloroplasts: all plants and photosynthetic microbes are descended from them as endosymbionts. Only cyanobacteria and the subsequent lineages (with both photosystems) use oxygen as the terminal electron acceptor in photosynthesis, meaning that they can produce oxygen. Cyanobacteria were responsible for the Great Oxidation Event.
So okay, it stands to reason that cyanobacteria acquired the photosystems from Somewhere else. The questions are: 1) When 2) From what organisms 3) Vertical or horizontal acquisition?
The most commonly accepted model right now is the “Fusion model,” in which cyanobacteria acquired photosystems II and I from different, relatively distant photosynthetic organisms.
The article I linked above takes a molecular clock approach to these questions. For those unfamiliar: the molecular clock is a useful metric because of neutral evolution, the process by which synonymous mutations (mutations that don’t affect the protein sequence due to the degeneracy of the aa code) occur in genomes at a predictable rate. Molecular clocks are “calibrated” using fossils whose ages are known. We can use the molecular clock to help date evolutionary events and build phylogenies.
The authors of this paper used a maximum likelihood statistical approach to construct a phylogeny for the major groups of early photosynthetic bacteria. The sequence data they used came from ribosomal proteins. The end result looks like this:
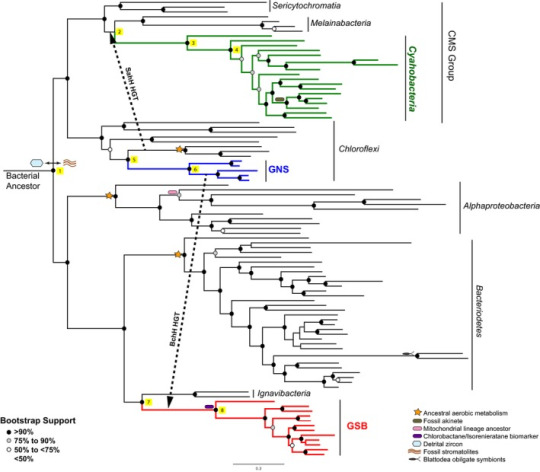
(Note: “Bootstrap support” is basically the percent chance that you’ve arrived at a particular branch/bifurcation nonrandomly. There's more to it than that, but I'm not gonna go into it here. That's the important bit.)
These authors assert that stem cyanobacteria likely received photosystem II genes via HGT from green non-sulphur bacteria (as seen above) prior to the Great Oxidation Event. They don’t think that something similar happened with photosystem I genes, however. Maybe those arose from within cyanobacteria themselves? Or a now extinct microbial line? Something else? It’s an open question.
It’s also worth pointing out that eukaryotic plant cells arose as a result of endosymbiosis with cyanobacteria. HOWEVER, this was also likely really convoluted. I’m not gonna go into this part in great detail, but there were at least five (but probably more like a dozen) separate endosymbiotic uptakes, leading to primary plastids. Additional uptakes of red and green algae led to secondary plastids. It’s awesome.
Okay! Whew. It’s been a little while since I took this class, so I hope all of that is right. Do check out the paper I linked, if you want, particularly if you’re a stats person.
And like. This is so cool you guys.
“God of the Gaps” is a logical fallacy in which people use things that science can’t yet explain as proof of God’s existence. I’ve heard people use God this way to try to explain these areas in evolutionary history where open questions still exist. How did abiogenesis, life from non-living matter, occur? We’ve got a lot of compelling biochemistry, but since there isn’t one simple answer, I’ve seen people handwave it away and just say God. More relevant here, how did oxidative photosynthesis evolve? There are still a lot of questions, and I do believe that they are questions that we can eventually, reasonably hope to answer.
What I really love is to see God IN the gaps. I love these sort of transitional, liminal phases in life’s history where we can see modern forms beginning to take shape. “Endless forms most beautiful,” Darwin wrote, just as God said “It is good;” but what about when “it” wasn’t exactly assembled yet? What about the time before the Great Oxidation Event when the stem cyanobacteria had just one photosystem?
That, I think, is where I see God hovering over the face of the deep. In deep time, I see him present with creation, the only one there to observe as genes that would oxidize our planet were transferred from one microbe to another, somewhere there in the deep.
#i need a tag for weekly science rambles#how about#endless forms most beautiful#i'll go back and tag the other one in a bit#block it if you're not interested#all truth is God's truth#pontifications and creations
4 notes
·
View notes
Text
Scientists in Australia made an incredible reproductive discovery: 17-million-year-old sperm. The male sex cell was found in the fossilized remains of freshwater crustaceans, known as ostracods.
The sperm themselves are an extraordinary find. They were thought to be longer than the male shrimplike creature's body, but were tightly coiled up inside the sexual organs.
. . .
“The Riversleigh fossil deposits in remote northwestern Queensland have been the site of the discovery of many extraordinary prehistoric Australian animals, such as giant, toothed platypuses and flesh-eating kangaroos," Archer said in a statement from UNSW. "So we have become used to delightfully unexpected surprises in what turns up there."
. . .
Just as bizarre as the discovery is how scientists think the fossil was preserved: bat poop.
Professor Archer explains: “About 17 million years ago, Bitesantennary Site was a cave in the middle of a vast biologically diverse rainforest. Tiny ostracods thrived in a pool of water in the cave that was continually enriched by the droppings of thousands of bats.
"The phosphorus from the stream of bat feces mixed with the water and helped preserve these fossils. Bat droppings have also helped preserve fossils in France.
----
This is an old article, but I thought it might be new and interesting to other people to.
Ostracods, or ostracodes, are a class of the Crustacea (class Ostracoda), sometimes known as seed shrimp. Some 70,000 species (only 13,000 of which are extant) have been identified,[1] grouped into several orders. They are small crustaceans, typically around 1 mm (0.039 in) in size, but varying from 0.2 to 30 mm (0.008 to 1.181 in) in the case of Gigantocypris. Their bodies are flattened from side to side and protected by a bivalve-like, chitinous or calcareous valve or "shell". The hinge of the two valves is in the upper (dorsal) region of the body. Ostracods are grouped together based on gross morphology. While early work indicated the group may not be monophyletic[2] and early molecular phylogeny was ambiguous on this front,[3] recent combined analyses of molecular and morphological data found support for monophyly in analyses with broadest taxon sampling.[4]

1 note
·
View note
Text
Blast (N/A) - What is BLAST?
Blast (N/A) aims to enter the Ethereum Layer 2 marketplace with a strong competitive edge. The network will distribute native yields of ETH and USDC via rebasing to dApps, offering higher returns than many other L2 solutions. However, the project has attracted a large number of skeptics, who call it a pyramid scheme or even a scam.
BLAST stands for Basic Local Alignment Search Tool and is a sequence similarity search algorithm. It compares a protein or nucleotide query sequence with a database of nucleotide or protein sequences to find matches that have significant similarities. Its use as a computational tool has revolutionized genomics and proteomics, forensic DNA analysis, drug discovery, and molecular medicine.
The BLAST program is typically run on several computers at the same time, utilizing parallel processing and multithreaded execution to speed up the search process. It also uses a memory hierarchy, which groups small and large structures into levels of cache, to reduce latency. It is one of the most widely used bioinformatics tools in the world.
The initial setup phase involves setting up a "lookup table" that maps words from the database to positions in the subject sequence. Then, during the scanning phase, each subject sequence is compared to the query for matches. High-scoring matches are saved for further processing. The program then generates gap-free or gapped Smith-Waterman local alignments for each of the initially found matches, if the E-value for each match is low enough.
Two of the largest structures in BLAST memory are the lookup table and the diag array, which tracks how far word hits have been extended on each diagonal. The lookup table has a size that grows as the query length increases. Both of these structures are frequently accessed, causing a significant part of the overall search time.
Other settings can influence the performance of a BLAST search, such as the default wordsize and substitution matrix. Changing these can improve the sensitivity of the search. Moreover, the BLAST program can be run with different algorithms to achieve specific types of results.
BLAST is a popular software package that can be extended to perform different bioinformatics tasks, such as detecting conserved domains, determining protein sub-cellular localization, and establishing phylogeny. It can also be used as an annotation tool for genome sequencing.
Blast is backed by notable investors like Paradigm and Standard Crypto, and is scheduled to launch in May 2024. The project has already attracted a considerable amount of funds and participants. The community has responded to the announcement with a wave of enthusiasm and excitement, but some have voiced concern that Blast is a pyramid scheme or scam. Pacman, the pseudonymous co-founder of NFT marketplace Blur and one of the project's main backers, has called such skepticism "unfair." Despite such doubts, Blast appears poised to gain traction in the L2 space.
0 notes