#rna
Text
Octopuses are curious and clever. They can solve mazes and puzzles, use tools, and are masters of camouflage. These complex abilities are powered by their sophisticated and giant brains.
Now, in the journal Cell, researchers report that octopuses are able to edit genetic information to quickly resculpt those brains when confronted with changes in their environment.
These findings cast new light on the incredible adaptability of these shape-shifting creatures and may help scientists design therapeutics for problematic mutations in our own bodies.
Continue Reading
435 notes
·
View notes
Photo


Punk-rock money cat for my little brother's 30th birthday
I won't point out why each piece of this outfit is relevant to him, but I think it's pretty explanatory. He is actually the one who took those pictures while we were waiting for the guests (which explains the lack of seriousness).
Outfit rundown
T-shirt: second-hand RNA
Leather jacket: thrifted
Vinyl pants: JJXX clearance bin
Shoes: Yosuke
Suspenders: second-hand Fint
Brooches and earring: thrifted/Design Festa/gifts
I had rings at one point, but took them off to help with the food and forgot to put them back
#fashion#jfashion#punk fashion#punk style#punk rock#alternative fashion#alt fashion#long hairstyles#second-hand fashion#leather jacket#money cat#rna#goth#goth fashion#goth style#gothic fashion#fanny rosie#fannyrosie
386 notes
·
View notes
Link
Researchers have introduced a groundbreaking computational framework called DEMINING, designed to directly identify expressed DNA and RNA mutations in RNA deep sequencing data. This innovative approach incorporates a deep learning model named DeepDDR, which effectively distinguishes expressed DNA mutations from RNA mutations following RNA-seq read mapping and pileup. Upon application to RNA-seq data from acute myeloid leukemia patients, DEMINING revealed previously overlooked DNA and RNA mutations, some of which were linked to the increased expression of host genes or the generation of neoantigens. Additionally, the study demonstrated DEMINING’s ability to accurately classify DNA and RNA mutations in RNA-seq data from non-primate species using transfer learning techniques. This development holds great promise for advancing our understanding of genetic mutations and their implications in various biological contexts.
Changes in DNA and RNA play significant roles in the domains of genetics evolution and human well-being. These changes determine the hereditary variety, considering species’ variation and development over the long run. Changes can create genetic issues and illness if they happen in basic and administrative domains. Cancerous growth can also usually be caused by DNA alterations in which oncogenes are activated or deactivated cancer silencer properties. These changes are critical in biodiversity and natural specializations, allowing species to thrive and evolve.
These changes in DNA can be frequently recognized using whole genome sequencing or from RNA-seq data; however, RNA mutations, such as Adenosine-to-Inosine (A-to-I) RNA changes, are difficult to differentiate because of the existence of expressed DNA Mutations. DEMINING, a stepwise bioinformatic technique, was developed to overcome this. DEMINING combines DeepDDR, a sophisticated learning model that distinguishes transmitted DMs from RMs in RNA-seq data. Even working with intense myeloid leukemia (AML) patients, DEMINING uncovered various ignored variations and changes in AML-related gene regions, revealing insight into potential diagnostic and therapeutic targets. This tool provides a new way of recognizing infection-related changes from RNA-seq datasets.
Continue Reading
41 notes
·
View notes
Text
Vribozyme, the RNA Pokémon!
Type: Poison
Evolves into ???
Being so simplistic, Vribozymes survive by drawing electrons and other forms of energy from nearby molecules. The red gem in the center is said to hold the RNA it’s composed of together, emanating a sort of glow that seems to resemble conscious thought.
#fakemon#pokemon#carbohs#natural history#paleontology#paleoblr#paleoart#evolution#RNA#ribozyme#sword
20 notes
·
View notes
Text

Back to working on these, I was planning on doing these in batches but I think it would just be easier to do one at a time. So here's RNA's version of Wildvine, or just a Florauna really
16 notes
·
View notes
Text
🔻-(Tw:eye-strain)-🔺

🐏-[Sybolisim for bone problems I have to try and explain how it feels]-🐏
#my art shit#sheep art#my artwork#sheep girl#my art#furry sheep#my artwrok#sheep#sheep fursona#red art#sheep furry#eye strain#disabled#disability#sheep sona#disabilties#red theme#bone problems#rna#chronic pain#bone pain#sheep oc#oc character#oc art#ocs#oc#disability problems
28 notes
·
View notes
Text
let's try this... without any coordination this time.
(honor system but pretty please just guess and dont tell anyone the current results/what to vote for; i want to see how much of a dumpster fire this becomes when left to its own devices)
126 notes
·
View notes
Photo
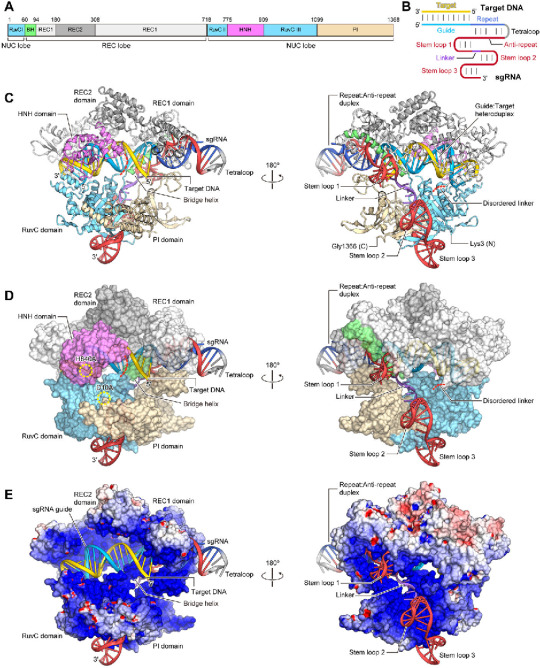
Figure 1. Overall structure of the Cas9–sgRNA–DNA ternary complex (A) Domain organization of S. pyogenes Cas9. BH, Bridge helix. (B) Schematic representation of the sgRNA:target DNA complex. (C) Ribbon representation of the Cas9–sgRNA–DNA complex. Disordered linkers are shown as red dotted lines. (D) Surface representation of the Cas9–sgRNA–DNA complex. The active sites of the RuvC (D10A) and HNH (H840A) domains are indicated by dashed yellow circles. (E) Electrostatic surface potential of Cas9. The HNH domain is omitted for clarity. Molecular graphic images were prepared using CueMol (http://www.cuemol.org).
Nishimasu, Hiroshi et al. “Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA.” Cell 156 (2014): 935-949.
137 notes
·
View notes
Text
My favorite thing about Homestuck is that all the chumhandles have initials made up of G, C, A and T, the four "letters" that make up DNA base pairs.
The notable exceptions are John's ectoBiologist handle, (justified because he was originally ghostyTrickster but changed it) and the cherubs, whose handles use the letter U. This is clever because uracil (U) replaces thymine (T) in RNA.
Considering the importance of DNA to the story, and the presence of a guardian named G-CAT, you'd think this would be something the fandom would discuss more or more frequently carry over to OC chumhandles.
27 notes
·
View notes
Text

Despite Bright Light
New approach that enables ten RNA and/or protein targets in whole zebrafish embryos and mouse brain sections to be visualised and quantified simultaneously, even in highly autofluorescence (natural emission of light by biological structures) samples
Read the published research article here
Image adapted from figure in work by Samuel J. Schulte and colleagues
Division of Biology & Biological Engineering, California Institute of Technology, Pasadena, CA, USA
Image originally published with a Creative Commons Attribution 4.0 International (CC BY 4.0)
Published in Development, February 2024
You can also follow BPoD on Instagram, Twitter and Facebook
8 notes
·
View notes
Text

Exploring RNA Interference
Imagine a molecular switch within your cells, one that can selectively turn off the production of specific proteins. This isn't science fiction; it's the power of RNA interference (RNAi), a groundbreaking biological process that has revolutionized our understanding of gene expression and holds immense potential for medicine and beyond.
The discovery of RNAi, like many scientific breakthroughs, was serendipitous. In the 1990s, Andrew Fire and Craig Mello were studying gene expression in the humble roundworm, Caenorhabditis elegans (a tiny worm). While injecting worms with DNA to study a specific gene, they observed an unexpected silencing effect - not just in the injected cells, but throughout the organism. This puzzling phenomenon, initially named "co-suppression," was later recognized as a universal mechanism: RNAi.
Their groundbreaking work, awarded the Nobel Prize in 2006, sparked a scientific revolution. Researchers delved deeper, unveiling the intricate choreography of RNAi. Double-stranded RNA molecules, the key players, bind to a protein complex called RISC (RNA-induced silencing complex). RISC, equipped with an "Argonaut" enzyme, acts as a molecular matchmaker, pairing the incoming RNA with its target messenger RNA (mRNA) - the blueprint for protein production. This recognition triggers the cleavage of the target mRNA, effectively silencing the corresponding gene.
So, how exactly does RNAi silence genes? Imagine a bustling factory where DNA blueprints are used to build protein machines. RNAi acts like a tiny conductor, wielding double-stranded RNA molecules as batons. These batons bind to specific messenger RNA (mRNA) molecules, the blueprints for proteins. Now comes the clever part: with the mRNA "marked," special molecular machines chop it up, effectively preventing protein production. This targeted silencing allows scientists to turn down the volume of specific genes, observing the resulting effects and understanding their roles in health and disease.
The intricate dance of RNAi involves several key players:dsRNA: The conductor, a long molecule with two complementary strands. Dicer: The technician, an enzyme that chops dsRNA into small interfering RNAs (siRNAs), about 20-25 nucleotides long. RNA-induced silencing complex (RISC): The ensemble, containing Argonaute proteins and the siRNA. Target mRNA: The specific "instrument" to be silenced, carrying the genetic instructions for protein synthesis.
The siRNA within RISC identifies and binds to the complementary sequence on the target mRNA. This binding triggers either:Direct cleavage: Argonaute acts like a molecular scissors, severing the mRNA, preventing protein production. Translation inhibition: RISC recruits other proteins that block ribosomes from translating the mRNA into a protein.
From Labs to Life: The Diverse Applications of RNAi
The ability to silence genes with high specificity unlocks various applications across different fields:
Unlocking Gene Function: Researchers use RNAi to study gene function in various organisms, from model systems like fruit flies to complex human cells. Silencing specific genes reveals their roles in development, disease, and other biological processes.
Therapeutic Potential: RNAi holds immense promise for treating various diseases. siRNA-based drugs are being developed to target genes involved in cancer, viral infections, neurodegenerative diseases, and more. Several clinical trials are underway, showcasing the potential for personalized medicine.
Crop Improvement: In agriculture, RNAi offers sustainable solutions for pest control and crop development. Silencing genes in insects can create pest-resistant crops, while altering plant genes can improve yield, nutritional value, and stress tolerance.
Beyond the Obvious: RNAi applications extend beyond these core areas. It's being explored for gene therapy, stem cell research, and functional genomics, pushing the boundaries of scientific exploration.
Despite its exciting potential, RNAi raises ethical concerns. Off-target effects, unintended silencing of non-target genes, and potential environmental risks need careful consideration. Open and responsible research, coupled with public discourse, is crucial to ensure we harness this powerful tool for good.
RNAi, a testament to biological elegance, has revolutionized our understanding of gene regulation and holds immense potential for transforming various fields. As advancements continue, the future of RNAi seems bright, promising to silence not just genes, but also diseases, food insecurity, and limitations in scientific exploration. The symphony of life, once thought unchangeable, now echoes with the possibility of fine-tuning its notes, thanks to the power of RNA interference.
#science sculpt#life science#science#molecular biology#biology#biotechnology#dna#double helix#genetics#artists on tumblr#rna#rna sequencing#RNA interference#cell biology#cells#biomolecules#illustrates#scientific illustration#illustration#illustrative art#scientific research
10 notes
·
View notes
Text
The Tasmanian tiger has a long, checkered history that now includes being the first extinct animal family from which scientists have recovered RNA, the molecule that brings a species' genome to life.
With a specially modified protocol, a Swedish team extracted millions of RNA strands from the skin and muscle of a 132-year-old Tasmanian tiger, or thylacine.
Known to scientists as Thylacinus cynocephalus, the striped, carnivorous marsupial was hunted to extinction on the island of Tasmania in Australia in the 1930s.
Recovering RNA – the genetic material that translates the information encoded in DNA into proteins – from extinct animals like the thylacine could open a trove of information on gene activity long thought lost.
Continue Reading.
164 notes
·
View notes
Link
AlphaFold’s breakthrough in solving the protein structure prediction problem has ignited enthusiasm to apply similar methods to predict the 3D structures of RNAs. However, achieving this feat presents significant challenges. A review paper titled ‘When will RNA get its AlphaFold moment?’ recently published in Nucleic Acids Research, discusses, in-depth, the various challenges faced by scientists over the years while attempting to develop deep learning models similar to AlphaFold for studying and predicting 3D structures of RNA.
A variety of techniques, ranging from physics to machine learning-based methods, have been employed to predict RNA structures. The studies stressed the fact that data-hungry deep learning methods are heavily reliant on large amounts of data, and in RNA structure prediction, the limited availability of structure and alignment data poses a significant challenge. Sequence data has also been found to be biased, and a lot of it does not uphold quality standards. Addressing and solving these issues is crucial to enabling further research into RNA prediction and opening new avenues.
Continue Reading
34 notes
·
View notes
Text

The first ancestor | O primeiro ancestral
🇬🇧
The origin of life on Earth is a fascinating and complex topic that continues to be the subject of scientific investigation and debate. The exact details of how and when life first emerged are not yet fully understood, but there are several theories and hypotheses.
One prominent theory suggests that life on Earth originated in hydrothermal vents at the ocean floor. These environments provided a rich mix of chemicals and minerals that could have supported the formation of simple organic molecules. Over time, these molecules might have developed into more complex structures, leading to the first self-replicating entities. Another hypothesis proposes that life's “building blocks” may have been delivered to Earth through comets or asteroids, carrying organic molecules from space. This concept, known as panspermia, suggests that the raw materials for life could have arrived on Earth from extraterrestrial sources.
The transition from simple organic molecules to the first living organisms is a critical step in the emergence of life. The earliest life forms were likely simple, single-celled organisms. These early life forms, often referred to as prokaryotes, lacked a distinct nucleus and membrane-bound organelles. They were likely similar to modern-day bacteria and archaea. One crucial aspect of early life was the development of a self-replicating system, allowing for the transmission of genetic information from one generation to the next. The formation of simple genetic material, possibly in the form of RNA, played a pivotal role in this process.
The study of extremophiles beings, organisms that thrive in extreme environments such as hot springs and deep-sea hydrothermal vents, has also provided valuable data into the conditions that early life might have faced. While the details remain a subject of ongoing research, understanding the origins of life on Earth is crucial for unraveling the broader story of our planet's history and the potential for life elsewhere in the universe. Scientists continue to explore these questions through various disciplines, including paleontology, molecular biology, and astrobiology.
🇧🇷
A origem da vida na Terra é um tópico fascinante e complexo que continua a ser objeto de investigação científica e debate. Os detalhes exatos de como e quando a vida surgiu ainda não são completamente compreendidos, mas existem várias teorias e hipóteses.
Uma teoria proeminente sugere que a vida na Terra teve origem em fontes hidrotermais no fundo do oceano. Esses ambientes forneciam uma mistura rica de produtos químicos e minerais que poderiam ter apoiado a formação de moléculas orgânicas simples. Com o tempo, essas moléculas poderiam ter se desenvolvido em estruturas mais complexas, levando aos primeiros seres auto-replicantes. Outra hipótese propõe que os “blocos de construção da vida” podem ter sido entregues à Terra por meio de cometas ou asteroides, carregando moléculas orgânicas do espaço. Esse conceito, conhecido como panspermia, sugere que os materiais para a vida podem ter chegado à Terra a partir de fontes extraterrestres.
A transição de moléculas orgânicas simples para os primeiros organismos vivos é um passo crítico no surgimento da vida. Os primeiros seres vivos provavelmente eram organismos unicelulares simples. Essas formas de vida iniciais, frequentemente chamadas de procariontes, não possuíam um núcleo distinto e organelas membranosas. Eles eram provavelmente semelhantes às bactérias e arqueias modernas. Um aspecto crucial da vida inicial foi o desenvolvimento de um sistema autorreplicante, permitindo a transmissão de informações genéticas de uma geração para outra. A formação de material genético simples, possivelmente na forma de RNA, desempenhou um papel fundamental nesse processo.
O estudo de seres extremófilos, organismos que prosperam em ambientes extremos como nascentes termais e fontes hidrotermais profundas, também forneceu insights valiosos sobre as condições que a vida inicial pode ter enfrentado. Embora os detalhes permaneçam um tema de pesquisa em andamento, compreender as origens da vida na Terra é crucial para desvendar a história mais ampla de nosso planeta e o potencial de vida em outros lugares do universo. Cientistas continuam a explorar essas questões por meio de várias disciplinas, incluindo paleontologia, biologia molecular e astrobiologia.
#space#universe#paleobotany#paleontology#science#digital painting#dinosaur#life#geology#biology#biodiversity#molecular biology#dna#rna
6 notes
·
View notes
Text

With Star, unlike RNA, they actually name their aliens, so get really for new dumb names.
The Splicer picked out their playlist for the purpose of forcing them to adapt and get creative, sink or swim.
They have RNA's thing of sticking close to their body type.
7 notes
·
View notes
Text
mRNA Translation
-- process of translating sequence of messenger RNA
-- messenger RNA = mRNA
-- turns sequence into amino acids
-- ribosome reads mRNA sequence in groups of three
-- three mRNA letters = codon
-- codons code for an amino acid
-- information begins in the DNA
-- transcribed into messenger RNA
-- translated from messenger RNA to protein
-- information is being transferred from one form to another
-- ribosome travels along the mRNA
-- detects the code as it goes
-- decides what amino acid to add based on the code
-- adds that amino acid to the polypeptide chain
-- this polypeptide chain becomes the protein
.
Patreon
#studyblr#notes#biology#bio#bio notes#biology notes#genetics#genetics notes#transcription#translation#mrna#messenger rna#rna#ribonucleic acid#dna#deoxyribonucleic acid#polypeptides#proteins#making proteins#life science#science#health science#medblr#medical notes#med notes#biochemistry#biochem#biochemistry notes#biochem notes#polypeptide chains
9 notes
·
View notes