#bioIT
Text
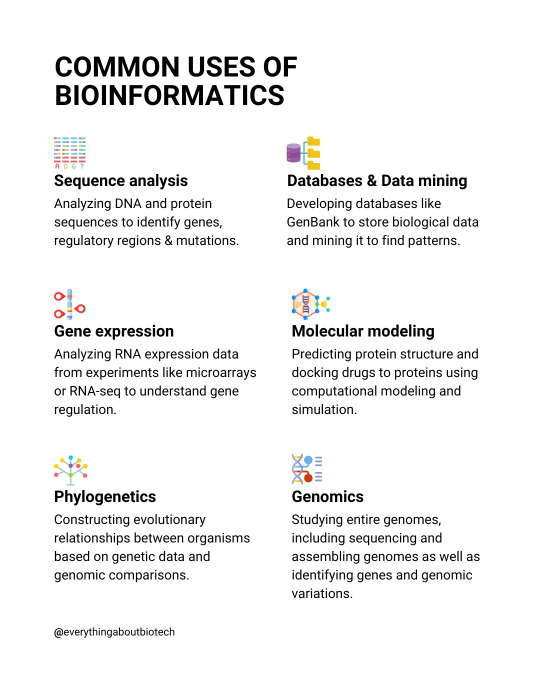
Common uses of bioinformatics
💡Sequence analysis
Analyzing DNA and protein sequences to identify genes, regulatory regions & mutations.
💡Gene expression
Analyzing RNA expression data from experiments like microarrays or RNA-seq to understand gene regulation.
💡Phylogenetics
Constructing evolutionary relationships between organisms based on genetic data and genomic comparisons.
💡Molecular modeling
Predicting protein structure and docking drugs to proteins using computational modeling and simulation.
💡Databases & Data mining
Developing databases like GenBank to store biological data and mining it to find patterns.
💡Genomics
Studying entire genomes, including sequencing and assembling genomes as well as identifying genes and genomic variations.
Follow @everythingaboutbiotech for useful posts.
#bioinformatics#genomics#proteomics#sequencing#PCR#biodata#bioIT#precisionmedicine#digitalhealth#biotech#DNA#healthtech#medtech#biostatistics#bioinformaticsjobs#BLAST#microarray#GenBank
34 notes
·
View notes
Text
BioIT World Conference Exhibitor List 2024

The Bio-IT World Conference 2024 is the world’s premier event showcasing the future of precision medicine. Connect with professionals and experts who are revolutionizing healthcare with our Bio-IT World Conference Exhibitor List 2024!
0 notes
Text
Elucidata at Bio-IT World Conference 2024: Accelerating Drug Discovery through AI-Ready Biomedical Data
BioIT World Conference and Expo, held on April 24th in Boston, is one of the biggest conferences that brings in professionals from the biomedical, bioinformatics, and IT sectors to converge, share insights, and explore the advancements shaping the future of Life Sciences. Like every other year, Elucidata attended the conference with a lot of zeal and enthusiasm. Here we talk about the highlights of the conference and our key takeaways.
Source Link
0 notes
Text

BioIT World Conference Exhibitor List 2024
Bio-IT World Conference 2024 event has united a community of leading life sciences, pharmaceutical, clinical, and healthcare together. Connect with industry pioneers with our Bio-IT World Conference Exhibitor List 2024!
0 notes
Text
TUMBLR YOU BITCH ZTOP SHOWING ME BLAZED POSTS ON MY FOLLOWING FEED !990 I LITRRSLLY DONT CARE ABOUT THAT SHIT IM HERE FOR FAN ART AND SHITTY TAKES OKAY !!! NOT PHOTOGRAPHY AND RANDOM SPORTS SHIT!!!! I AM HERE FOR MY FRIENDS NOT FOR YOUR FUCKING PROFIT BIOIT.
1 note
·
View note
Text
Structure-guided Drug Discovery: (1) The Coronavirus 3CL hydrolase (Mpro) enzyme (main protease) essential for proteolytic maturation of the virus and (2) viral protease, the RNA polymerase, the viral spike protein, a viral RNA as promising two targets for discovery of cleavage inhibitors of the viral spike polyprotein preventing the Coronavirus Virion the spread of infection

Structure-guided Drug Discovery: (1) The Coronavirus 3CL hydrolase (Mpro) enzyme (main protease) essential for proteolytic maturation of the virus and (2) viral protease, the RNA polymerase, the viral spike protein, a viral RNA as promising two targets for discovery of cleavage inhibitors of the viral spike polyprotein preventing the Coronavirus Virion the spread of infection
Curators and…
View On WordPress
#BioIT: BioInformatics#Biological Engineering#Biological Networks#Biomarkers & Medical Diagnostics#Cancer Informatics#Clinical & Translational#clinical genomics#Commercialization#COVID-19#Gene Regulation and Evolution#Genetics & Pharmaceutical#Immunotherapy#Innovation in Immunology Diagnostics#Innovations#Intellectual property#Investment in Technological Breakthrough#NGS#Pharmaceutical R&D Informatics#Population Health Management#Proteomics#SARS-CoV#SARS-CoV-2#Universal Immune Cell Therapies (uICT)#Virus Infective Acute Respiratory Syndrome: SARS-CoV | Tagged COVID-19
0 notes
Note
okei oikeesti ootko aallos ja opiskeleks bioIT/LST/jotain chemin bio hommaa? t-teekkari?

:)
3 notes
·
View notes
Text
my youtube algorithm after I watch 65 programming videos in one day as a last desperate attempt to pass my bioIT exam

#also my yt algorhitm watching me spell 'algorhitm' incorrectly for the sixteenth time#yes bc im learning abt algorhitms#I Am So Tired#I havent passed an exam in months#personal#uni tag
10 notes
·
View notes
Text
So You Want to Start a Biotech: A Bioinformatics Approach That Works
In the middle of a raging pandemic, two small biotechs (BioNTech and Moderna) have stepped up to save the world.
Biotechs are awesome.
If you want to start a biotech, you’re going to need:
A great idea
A boatload of funding
A can-do attitude
Some sort of wet lab setup
Some sort of bioinformatics setup
I am here to help with that last one with some guiding principles and a tech stack that I have used at a couple of startups that basically worked.
I did not come up with the tech stack.
A version of it was originally set up by Andrew Clark who I worked with at Gritstone Oncology and learned a lot from. It is described from the dev ops perspective with more technical details here:
https://aws.amazon.com/partners/success/gritstone-oncology/
Here is the bioinformatics/biologist version:
1) Startups need to move quickly.
An investor funds a biotech with a lump of money and the expectation that the company will get from point A to point B. If you don’t get to point B, the investor isn’t going to give you any more money. If you run out of money before you get to point B, you are toast.
2) You don’t know what you are doing
The first stages of a biotech company are always research and development. The development part means that things keep changing.
Your tech stack should support getting you from point A to point B. It doesn’t necessarily need to support the final product yet. Your final product is a moving target and if you focus too much on getting that right and not getting to point B, you will burn a lot of your runway writing a lot of code that ultimately gets deleted (and by deleted I mean stuck in a GitHub repository never to be opened again).
3) Take on technical debt
Technical debt is the cost that you have to pay for putting off for tomorrow the stuff you could have done today. Normally, it’s cheaper to do stuff properly so you don’t have to revisit it.
In bioinformatics, and particularly in R&D, we can be a little flexible about doing things “properly.” You don’t have to pay technical debt on code that gets deleted. This isn’t an excuse to be sloppy. You should, for example, still comment your code.
I am just saying that many successful biotechs were built on shoddy Unix scripts.
We don’t have to go that far anymore, but if your programmers are swearing at you in five years about your terrible code, that’s a win! You lasted five years and grew enough to hire judgmental programmers! Congratulations!
These three points are actually the things I have seen people struggle getting their heads around. If you get right on these, the rest is just connecting the dots.
Here is the tech stack:
4) Stick everything on an Amazon cloud
Yeah, it costs a lot.
But cloud services give you simplicity and scalability that you can’t get from a stack of servers in a closet. This is important because you don’t know what you are doing yet so you don’t what you need.
There are a bunch of cloud solutions. I have used Amazon and that worked fine. A very basic setup would be as follows:
An s3 (simple storage service) bucket where you store most your data.
An EC2 (Elastic cloud compute) instance where your run your code
Easy, right?
The s3 buckets, if you haven’t used them, are a storage system. They look like a big computer but have a hokey and annoying file system that takes a little getting used to. They are good places to store large datasets, like the outputs of sequencing runs, but you don’t do computing on s3.
The EC2 instances look and act like regular computers/servers and can read and write data to the buckets. You can vary the size of your instance in the cloud so they can grow bigger or smaller based on how much compute power you need.
A better version of this would be to have two EC2 instances: One where you log in and one where you spin up nodes to run big jobs. These can be joined with a job scheduler such as UGER. This is a cheaper solution because you only pay for the nodes when you are using them. It also prevents two users from running into each other if they are trying to use the same resources. From here, there are many different ways to go, depending on your needs.
Amazon cloud services uses a pay-for-what-you-use model, so in general you get a bigger bill if you store more data and use more compute. The size of the bill is also based on how well you configure your setup, to make sure you aren’t reserving more things than you are using.
The exact configuration is something a good dev ops person can help you sort out.
Amazon has a biotech Quickstart guide here: https://aws.amazon.com/quickstart/biotech-blueprint/
5) Use Nextflow to run your pipelines
There are two types of biotechs: Ones that know they are biotechs and ones that think they are software development shops.
Don’t roll your own software for things where excellent solutions already exist. Especially for things that aren’t science and aren’t core to your business.
One of the biggest pain points in bioinformatics is pipelines. Pipelines are software that run other software.
For example, your complete RNA pipeline might run sequence alignment, transcript quantification, and a bunch of intermediary steps to get stuff in the right formats. Pipeline software glues all the steps together. Really great pipeline software glues it all together and makes it easy to develop and run.
Nextflow is a really great pipeline software solution. In short, it takes your shoddy Unix scripts and makes them into real code.
It takes a couple days to get used to it, but once you do it saves you a lot of time. It forces you to make your code modular with inputs and outputs. It handles multhreading on the cloud for you so you don’t have break things up into chunks or spin up multiprocessing yourself. And it can restart jobs once they fail so you can correct code and restart pipelines without, e.g. commenting out parts that already ran.
When I was at ReadCoor, I used Nextflow in my group from the beginning. It gave me the resources of a mature company from day one. For example, I set up a complete NGS stack in a couple weeks, which I could never have done with Unix scripts. And it’s free!
Nextflow also has a tower solution that controls scheduling for jobs that have to run over and over. I haven’t used that myself as it wasn’t something I needed, but for many it would be worth investigating.
There is good support within the community with the Nextflow Core project providing a lot of common bioinformatics pipelines already set up. The team also spun up a consulting service called Seqera Labs that can help you optimize your Nextflow setup (for a fee).
And if you say Nextflow three times in the mirror* on Twitter the developers will pop up and give you advice and moral support.
6) Write all your code in python
Yeah, yeah, yeah.
Everyone has a favorite language. Maybe yours isn’t python. I could generalize this should say, “choose a language and stick with it.” But I won’t. You should use python.
It is a good all-rounder in terms of functionality and scalability.
It’s really popular across domains. This means you can hire either straight up programmers or more specialized bioinformaticians who know it.
Python is inherently scalable so it can be used to code small things, like plotting scripts in Jupyter notebooks, or big collaborative projects with code bases and classes.
It is also a data sciences powerhouse. Pandas is like a domain language onto itself. Python has packages to do advanced statistics, machine learning, and smorgasbord of visualizations.
“All your code”, of course, should be taken with a grain of salt. Python can be slow and you might some day need to write some things in C++ or Rust or something.
That’s OK.
We’re just getting started.
7) Use GitHub for version control
Put your code in GitHub. I think everyone already knows this.
8) Write a specification now and then
It won’t kill you and it is good for setting expectations between wet and dry lab.
9) Install a great LIMS solution
Hahahahahahhaha! Good luck. [Bioinformatician hides under desk]
10) Be good people
Finally, bioinformatics is a small community of highly skilled people with big mouths and high standards.
You can be competitive and expect a lot from people, but if you cross the line from “good person” to “not so much” everyone is going to know.
Who’s going to do all of this stuff?
Sometimes if your needs are small this can all be outsourced. There are some really great consulting companies around now. A couple ones I know for bioinformatics are Diamond Age Data Sciences and Fulcrum Genomics. KindlyOps handles compliance issues well. BioTeam does BioIT.
But if your needs are larger, or will grow larger, it’s a good idea to at least have a roadmap in your head of where you are going.
If you want to get all the skills you will need in house, you almost always have to hire from the top down. That usually means hiring really skilled people who are going to have to do grunt work for a while until the teams are sorted. Make sure they are cool with this. (Explicitly. Ask during the interview.)
Your wet lab and your dry lab should be two arms of the same lab. They need to work together, from the experiment’s design to its finish. Make sure they are cool with this.
A good core team at the beginning might consist of a bioinformatics scientist and a developer who knows dev ops on the cloud really well. If they are going to have core responsibilities within the company, their experience, salary, and hiring level should reflect that.
Outsource your IT and desktop support. This is a different skill set and your developers should not be setting up printers. They will probably do it badly and the wet lab will get annoyed and you don’t need drama to erupt about your printers.
Consider having an experienced HR professional at the C suite level. Recruiting in biotech is a nightmare and outsourcing recruitment is really expensive with varying results. A good HR person will take care of that.
Also, every startup is at least a little bit toxic. It is just the reality of having a small team with big goals. An HR professional will do some of the very draining emotional labor of keeping the company culture afloat. That will improve your tech stack. Really.
Finally, you can’t properly talk about hiring in biotech without addressing diversity. We all have biases, and we cannot counteract them by merely trying to be a good person (though that’s certainly a start). There are concrete techniques that can help us hire the most talented team, like removing names from resumes before evaluating them. A good HR leader should be able to help you find training and establish policies to do this properly.
So that’s it.
This is far from the only approach that will work. It is just one that I have seen work. There are many paths to the top of the mountain.
Biology offers us a universe of problems and biotech offers a universe of solutions for making the world a healthier, safer place.
Don’t let the basics get in your way.
Good luck!
*Joke shamelessly stolen from Mick Watson
Edit: Because I don’t want to inadvertently slight anyone or upset the Snakemake fans, there are other pipeline tools besides Nextflow that are worth looking into. Also, many great in house solutions (l am learning to love Martian) live in slightly older biotechs and were written from necessity before the community pipeline tools became mature.
3 notes
·
View notes
Text
BioIT World Conference Exhibitor List 2024

The Bio-IT World Conference 2024 is the world’s premier event showcasing the future of precision medicine. Connect with professionals and experts who are revolutionizing healthcare with our Bio-IT World Conference Exhibitor List 2024!
0 notes
Text

BioIT World Conference Exhibitor List 2024
Bio-IT World Conference 2024 offers a multitude of opportunities for attendees to engage in educational sessions, panel discussions, and workshops led by industry luminaries.
0 notes
Text
DOWNLOAD KE850 USB DRIVER
Downloads: 5631
File Name: ke850 usb driver
Uploader: Chip
File Version: 113141461
Operating Systems: Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X
Date Added: 13 December, 2019
Price: Free
Download Type: http
File Format: exe
File Size: 23 Mb

Fixed Issues:
- SMB](ke850 usb driver SMB]) - Fixed a bug where a shared folder could not be accessed by a “guest” user.
- Fixed data corruption issue(ke850 usb driver issue) in current v1.03 f/w after changing the toolchain.
- Fixed Issues: - Rocket League may experience an application hang on performing a task switch.
- Fixed Start recording and plug-out SD(ke850 usb driver SD) Card, Recording icon don't disappered.
- Fixed the issue that the system may not be able to boot up from SiliconImage RAID controller when Intel RAID BIOS is enabled.
- System]: Fixed a bug where a working drive was erroneously identified as malfunctioning.
- Fixed(ke850 usb driver Fixed) an issue that Time zone setting for Minsk should be GMT+3.
- Fixed Issues:- Possible Genlock failure under heavy load.
- Fixed OS installed in the SATA hard disk when both the SATA and PATA hard disks are present ,the PATA hard disk contains the Windows boot files.
- Fixed multiple localization issue(ke850 usb driver issue) on Catalyst Control Center.
Users content:
Change the link location for D-Link support site. And, you can preview corrections on the built-in LCD before you print. Intel P43 ICH10 Chipsets. The Basic Input/Output System (BIOIt is highly recommended to always use the most recent driver version available. These files are not harmful to your computer. Insert USB storage into your VEOLO 4K player. America; S&C America NOTE: - This version is for use with version V1.5.1 or above of the EAP300 software. Supports USB keyboard of new spec. The Epson Status Monitor 3 is incorporated into this driver. Fix Wireless stop service when too much interference in user environment. Open the link if you need the DOWNLOAD SONY VAIO VGN-SZ660NUPDATE DRIVER.
Supported OS:
Windows XP 32-bit
Windows Server 2003 32-bit
Windows 8.1/8/7/Vista 64-bit
Microsoft Windows 8 Enterprise (32-bit)
Microsoft Windows 8 (32-bit)
Windows Server 2008 R2
Microsoft Windows 8 Pro (64-bit)
Windows 7 64-bit
Windows Server 2016
Microsoft Windows 8.1 (64-bit)
Microsoft Windows 8.1 Pro (64-bit)
Microsoft Windows 8 Enterprise (64-bit)
Microsoft Windows 10 (32-bit)
Microsoft Windows 8.1 Enterprise (32-bit)
Windows 8
Windows Vista 32-bit
Microsoft Windows 8.1 Enterprise (64-bit)
Windows 2000
Windows 8.1/8/7/Vista 32-bit
Windows Server 2008
Notebook 8.1/8/7 64-bit
Windows 7 32-bit
Microsoft Windows 8 (64-bit)
Windows 10
Microsoft Windows 8 Pro (32-bit)
Windows Server 2012
Windows 7
Microsoft Windows 8.1 (32-bit)
Windows 8.1
Windows XP 64-bit
Notebook 8.1/8/7 32-bit
Microsoft Windows 10 (64-bit)
Microsoft Windows 8.1 Pro (32-bit)
Windows Server 2012 R2
Windows Server 2003 64-bit
Windows Vista 64-bit
Searches:
ke850 usb Znx163-nxy; ke850 usb driver; ke850 usb Z16n; ke850 usb driver for Microsoft Windows 8 Enterprise (32-bit); ke850 usb Z ZW163-1; ke850 usb driver for Windows Server 2012 R2; ke850 usb ZW1634; ke850 usb ZWKVX1634; ke850 usb driver for Windows Server 2008 R2; ke850 usb driver for Windows 2000; ke850 usb driver for Microsoft Windows 8.1 (32-bit)
Compatible Devices:
Apple; Video Projector; Laptop; Macbook; Ipad; Videocard
To ensure the integrity of your download, please verify the checksum value.
MD5: e53e550b8d912aca3150118f3ca15797
SHA1: 06576a781f7852e7dcc8282a46cf924d3de1e5b8
SHA-256: b64efabf2484dae18983a17a72fd76ed27ff8564abd239c2244c5cd425900919
0 notes
Text
18th Annual 2019 BioIT, Conference & Expo, April 16-18, 2019, Boston, Seaport World Trade Center, Track 5 Next-Gen Sequencing Informatics - Advances in Large-Scale Computing
18th Annual 2019 BioIT, Conference & Expo, April 16-18, 2019, Boston, Seaport World Trade Center, Track 5 Next-Gen Sequencing Informatics – Advances in Large-Scale Computing
18th Annual 2019 BioIT, Conference & Expo, April 16-18, 2019, Boston, Seaport World Trade Center, Track 5 Next-Gen Sequencing Informatics – Advances in Large-Scale Computing
https://www.bio-itworldexpo.com/programs https://www.bio-itworldexpo.com/next-gen-sequencing-informatics
Leaders in Pharmaceutical Business Intelligence (LPBI) Group represented by Founder & Director, Aviva…
View On WordPress
0 notes
Photo

#BioIT #AI #BigData @CHI_Healthtech @Frost_Sullivan @FS_Healthcare http://bit.ly/2waxBR3 pic.twitter.com/cUo5Q9YfzY #BioIT #AI #BigData @CHI_Healthtech @Frost_Sullivan @FS_Healthcare pic.twitter.com/cUo5Q9YfzY source
0 notes