#shotgun metagenomics
Photo

A new resource has been released that enables the comparison of microbial 16S rRNA and shotgun metagenomics data. The resource is a web-based tool that allows users to upload and compare two datasets, and to visualize the results as a heat map. The tool is designed to help researchers identify microorganisms that are present in different samples.
#Featured#News#OMICs#Topics#Translational Medicine#fault#microbial 16S rRNA#shotgun metagenomics#web-based tool#compare datasets#visualize results
0 notes
Text
Metagenomics and Metatranscriptomics: New Insights and Pipelines to Better Navigate Data Analysis

Scientists at the Institute of Parasitology and Biomedicine and the University of Granada, Spain, along with collaborators, developed two pipelines that could automate and optimize metagenomics and metatranscriptomics data analysis. These pipelines could be adapted for 16S, shotgun, and RNA-Seq data. Its performance was validated through three studies by assessing its taxonomy classification ability.
When Anton van Leeuwenhoek first opened the doors to the unseen world of microorganisms in 1673 through his self-made single-lens microscope, it couldn’t have been possible to imagine the explosion of discoveries that were to follow in its wake. The paradoxical world of microbes is a source of infinite curiosity to many scientists around the world. Thus, it was a no-brainer that with the advent of NGS, the microbes would get their very own niche within it—Metagenomics.
Continue Reading
62 notes
·
View notes
Text
You see, my problem with genetics and DNA is that it's like having this huge elaborate program, much like, let's say Sims 4, that's written in just binary code. A corrupted binary code may I add, it's not clearly available beginning to end, no. You have to make multiple copies of the binary string and cut it in smaller strings and then compare them to other small strings and see where they overlap, to try to create a longer chain. I'm not kidding, it's how shotgun metagenomics work.
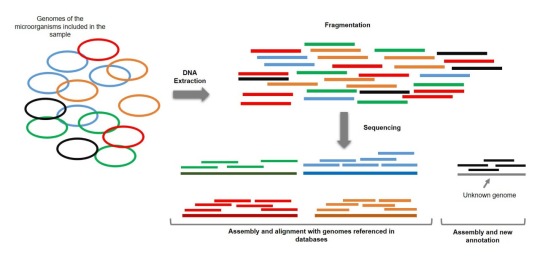
And it's not just crucial code, some parts are just programmers comments or previous test versions, or straight up gibberish.
I feel like there should be a much efficient way of doing things. Like a programming language equivalent, much like python or r or whatever, that we could access.
6 notes
·
View notes
Link
0 notes
Text
Extensive novel diversity and phenotypic associations in the dromedary camel microbiome are revealed through deep metagenomics and machine learning
The dromedary camel, also known as one-humped camel or Arabian camel, is an iconic and economically important feature of Arabian society. Its contemporary importance in commerce and transportation, along with the historical and modern use of its milk and meat products for dietary health and wellness, make it an ideal subject of scientific scrutiny. The gut microbiome has recently been associated with numerous aspects of health, diet, lifestyle, and disease in livestock and humans alike, as well as serving as an exploratory and diagnostic marker of many physical characteristics. Our initial pilot analysis of 55 camel gut microbiomes from the Fathi Camel Microbiome Project using deep metagenomic shotgun sequencing reveals substantial novel species-level microbial diversity, for which we have generated an extensive catalog of prokaryotic metagenome-assembled microorganisms (MAGs) as a foundational microbial reference database for future comparative analysis. Exploratory correlation analysis exhibits substantial correlation structure among the collected subject-level metadata, including physical characteristics. Machine learning using these novel microbial markers, as well as statistical testing, demonstrates strong predictive performance of microbes to distinguish between multiple dietary and lifestyle characteristics in dromedary camels. We derive strongly predictive models for camel age, diet (including wheat), level of captivity. These findings and resources represent substantial strides toward understanding the camel microbiome and pave the way for a deeper understanding of the camel microbiome, as well as the nuanced factors that shape camel health. http://dlvr.it/T3BJnD
0 notes
Text
Comparison of Blood-Based Shotgun and Targeted Metagenomic Sequencing for Microbiological Diagnosis of Infective Endocarditis
Pubmed: http://dlvr.it/T05Jnh
0 notes
Text
Search algorithm reveals nearly 200 new kinds of CRISPR systems
New Post has been published on https://thedigitalinsider.com/search-algorithm-reveals-nearly-200-new-kinds-of-crispr-systems/
Search algorithm reveals nearly 200 new kinds of CRISPR systems


Microbial sequence databases contain a wealth of information about enzymes and other molecules that could be adapted for biotechnology. But these databases have grown so large in recent years that they’ve become difficult to search efficiently for enzymes of interest.
Now, scientists at the McGovern Institute for Brain Research at MIT, the Broad Institute of MIT and Harvard, and the National Center for Biotechnology Information (NCBI) at the National Institutes of Health have developed a new search algorithm that has identified 188 kinds of new rare CRISPR systems in bacterial genomes, encompassing thousands of individual systems. The work appears today in Science.
The algorithm, which comes from the lab of pioneering CRISPR researcher Professor Feng Zhang, uses big-data clustering approaches to rapidly search massive amounts of genomic data. The team used their algorithm, called Fast Locality-Sensitive Hashing-based clustering (FLSHclust) to mine three major public databases that contain data from a wide range of unusual bacteria, including ones found in coal mines, breweries, Antarctic lakes, and dog saliva. The scientists found a surprising number and diversity of CRISPR systems, including ones that could make edits to DNA in human cells, others that can target RNA, and many with a variety of other functions.
The new systems could potentially be harnessed to edit mammalian cells with fewer off-target effects than current Cas9 systems. They could also one day be used as diagnostics or serve as molecular records of activity inside cells.
The researchers say their search highlights an unprecedented level of diversity and flexibility of CRISPR and that there are likely many more rare systems yet to be discovered as databases continue to grow.
“Biodiversity is such a treasure trove, and as we continue to sequence more genomes and metagenomic samples, there is a growing need for better tools, like FLSHclust, to search that sequence space to find the molecular gems,” says Zhang, a co-senior author on the study and the James and Patricia Poitras Professor of Neuroscience at MIT with joint appointments in the departments of Brain and Cognitive Sciences and Biological Engineering. Zhang is also an investigator at the McGovern Institute for Brain Research at MIT, a core institute member at the Broad, and an investigator at the Howard Hughes Medical Institute. Eugene Koonin, a distinguished investigator at the NCBI, is co-senior author on the study as well.
Searching for CRISPR
CRISPR, which stands for clustered regularly interspaced short palindromic repeats, is a bacterial defense system that has been engineered into many tools for genome editing and diagnostics.
To mine databases of protein and nucleic acid sequences for novel CRISPR systems, the researchers developed an algorithm based on an approach borrowed from the big data community. This technique, called locality-sensitive hashing, clusters together objects that are similar but not exactly identical. Using this approach allowed the team to probe billions of protein and DNA sequences — from the NCBI, its Whole Genome Shotgun database, and the Joint Genome Institute — in weeks, whereas previous methods that look for identical objects would have taken months. They designed their algorithm to look for genes associated with CRISPR.
“This new algorithm allows us to parse through data in a time frame that’s short enough that we can actually recover results and make biological hypotheses,” says Soumya Kannan PhD ’23, who is a co-first author on the study. Kannan was a graduate student in Zhang’s lab when the study began and is currently a postdoc and Junior Fellow at Harvard University. Han Altae-Tran PhD ’23, a graduate student in Zhang’s lab during the study and currently a postdoc at the University of Washington, was the study’s other co-first author.
“This is a testament to what you can do when you improve on the methods for exploration and use as much data as possible,” says Altae-Tran. “It’s really exciting to be able to improve the scale at which we search.”
New systems
In their analysis, Altae-Tran, Kannan, and their colleagues noticed that the thousands of CRISPR systems they found fell into a few existing and many new categories. They studied several of the new systems in greater detail in the lab.
They found several new variants of known Type I CRISPR systems, which use a guide RNA that is 32 base pairs long rather than the 20-nucleotide guide of Cas9. Because of their longer guide RNAs, these Type I systems could potentially be used to develop more precise gene-editing technology that is less prone to off-target editing. Zhang’s team showed that two of these systems could make short edits in the DNA of human cells. And because these Type I systems are similar in size to CRISPR-Cas9, they could likely be delivered to cells in animals or humans using the same gene-delivery technologies being used today for CRISPR.
One of the Type I systems also showed “collateral activity” — broad degradation of nucleic acids after the CRISPR protein binds its target. Scientists have used similar systems to make infectious disease diagnostics such as SHERLOCK, a tool capable of rapidly sensing a single molecule of DNA or RNA. Zhang’s team thinks the new systems could be adapted for diagnostic technologies as well.
The researchers also uncovered new mechanisms of action for some Type IV CRISPR systems, and a Type VII system that precisely targets RNA, which could potentially be used in RNA editing. Other systems could potentially be used as recording tools — a molecular document of when a gene was expressed — or as sensors of specific activity in a living cell.
Mining data
The scientists say their algorithm could aid in the search for other biochemical systems. “This search algorithm could be used by anyone who wants to work with these large databases for studying how proteins evolve or discovering new genes,” Altae-Tran says.
The researchers add that their findings illustrate not only how diverse CRISPR systems are, but also that most are rare and only found in unusual bacteria. “Some of these microbial systems were exclusively found in water from coal mines,” Kannan says. “If someone hadn’t been interested in that, we may never have seen those systems. Broadening our sampling diversity is really important to continue expanding the diversity of what we can discover.”
This work was supported by the Howard Hughes Medical Institute; the K. Lisa Yang and Hock E. Tan Molecular Therapeutics Center at MIT; Broad Institute Programmable Therapeutics Gift Donors; The Pershing Square Foundation, William Ackman and Neri Oxman; James and Patricia Poitras; BT Charitable Foundation; Asness Family Foundation; Kenneth C. Griffin; the Phillips family; David Cheng; and Robert Metcalfe.
#acids#algorithm#Algorithms#Analysis#Animals#Antarctic#approach#Artificial Intelligence#Bacteria#Big Data#biodiversity#Biological engineering#Biology#biotechnology#Brain#Brain and cognitive sciences#brain research#Broad Institute#cell#Cells#clusters#coal#Community#Computational biology#Computer science and technology#CRISPR#data#Database#databases#defense
0 notes
Quote
The gut microbiome plays an important role in the immune system development, maintenance of normal health status, and in disease progression. In this study, we comparatively examined the fecal microbiomes of Amish (rural) and non-Amish (urban) infants and investigated how they could affect the mucosal immune maturation in germ-free piglets that were inoculated with the two types of infant fecal microbiota (IFM). Differences in microbiome diversity and structure were noted between the two types of fecal microbiotas. The fecal microbiota of the non-Amish (urban) infants had a greater relative abundance of Actinobacteria and Bacteroidetes phyla, while that of the Amish (rural) counterparts was dominated by Firmicutes. Amish infants had greater species richness compared with the non-Amish infants' microbiota. The fecal microbiotas of the Amish and the non-Amish infants were successfully transplanted into germ-free piglets, and the diversity and structure of the microbiota in the transplanted piglets remained similar at phylum level but not at the genus level. Principal coordinates analysis (PCoA) based on Weighted-UniFrac distance revealed distinct microbiota structure in the intestines of the transplanted piglets. Shotgun metagenomic analysis also revealed clear differences in functional diversity of fecal microbiome between Amish and non-Amish donors as well as microbiota transplanted piglets. Specific functional features were enriched in either of the microbiota transplanted piglet groups directly corresponding to the predominance of certain bacterial populations in their gut environment. Some of the colonized bacterial genera were correlated with the frequency of important lymphoid and myeloid immune cells in the ileal submucosa and mesenteric lymph nodes (MLN), both important for mucosal immune maturation. Overall, this study demonstrated that transplantation of diverse IFM into germ-free piglets largely recapitulates the differences in gut microbiota structure between rural (Amish) and urban (non-Amish) infants. Thus, fecal microbiota transplantation to germ-free piglets could be a useful large animal model system for elucidating the impact of gut microbiota on the mucosal immune system development. Future studies can focus on determining the additional advantages of the pig model over the rodent model.
Amish (Rural) vs. non-Amish (Urban) Infant Fecal Microbiotas Are Highly Diverse and Their Transplantation Lead to Differences in Mucosal Immune Maturation in a Humanized Germfree Piglet Model - PMC
0 notes
Text
Fwd: Graduate position: UBarcelona.GutMicrobiotaEvolutionInsularLizards
Begin forwarded message:
> From: [email protected]
> Subject: Graduate position: UBarcelona.GutMicrobiotaEvolutionInsularLizards
> Date: 1 September 2023 at 07:16:59 BST
> To: [email protected]
>
>
>
> A PhD position is available at the Department of Evolutionary Biology,
> Ecology and Environmental Sciences of the University of Barcelona (Spain)
> to study the “Coevolution of the symbiosis between host and the gut
> microbiota in insular lizards from the Balearic Islands (Spain)”. The
> project aims at combining multi-omics with individual and population-level
> data to understand the strength of the symbiosis over a short-evolutionary
> time frame (within and across populations of the same species) and the
> potential role of the gut microbiome in extending host fitness landscape
> in resource-limited environments (small islands).
>
> The contract is available for 4 years (full funded), starting in last
> trimester 2023 or beginning of 2024 and it is financed by the Agencia
> Estatal de Investigación under the recently funded project "
>
> The island syndrome: toward an integration of life history traits,
> gut symbionts and population dynamics (ISLAB)."
>
> The project will be a collaboration between the University of Barcelona
> (https://ift.tt/8MZPaVR) and the group of Giacomo Tavecchia at the
> Mediterranean Institute for Advanced Studies (IMEDEA) in Mallorca (Spain).
>
> We look for a highly motivated PhD student, with passion for microbes,
> ecology, evolution and multi-omics data analysis.
>
> The PhD candidate will be involved in:
>
> Field sampling in the Balearic Islands Molecular lab work for data
> production (16S full-length, shotgun metagenomics and metabolomics)Gut
> microbiota analyses and integration with host metadata at individual-level
> (sex, age, morphometrics, genetics) and population/island level (life
> history traits, dynamics, diet, ecology)
>
> Ideal candidates will have:
>
> A B.Sc. or M.Sc. degree in biology degree in a field of biological
> sciences, and some prior research experience on microbial analysis Research
> and theoretical proficiency in microbiology, ecology, evolutionary
> theory Research experience in molecular lab work and bioinformatics
> (familiar with Unix and R programming)A strong interest in
> interdisciplinary research, with a focus on biological questions
> rather than the particular model organism. Strong written, oral, and
> interpersonal communication skills.
>
> To apply, please send a cover letter with a CV and contact information for
> three references (phone and email) to prof. Laura Baldo [email protected]
>
>
> Laura Baldo, PhD
> Associate Professor
>
> (Profesora agregada)
>
> Department of Evolutionary Biology, Ecology and Environmental Sciences
> (BEECA),
>
> Institut de Recerca de la Biodiversitat,
>
> University of Barcelona
> Av. Diagonal, 643 (Margalef building),5th floor
> 08028 Barcelona, Spain
> email: [email protected]
> phone: (+34) 9340 37144
>
> https://ift.tt/5B8JfRC
>
> Web page:
> http://www.ub.edu/evok
>
>
> Laura Baldo
1 note
·
View note
Text
MarketsandMarkets Infectious Disease and Molecular Diagnostics Conference 2023: A Gateway to the Latest Innovations in Microbial Metagenomics for Cancer Patients

Introduction:
Cancer patients often face multiple challenges, including a compromised immune system and increased susceptibility to infections. Accurate and timely diagnosis of infectious diseases is crucial in managing cancer patients' health. In recent years, the field of microbial metagenomics has emerged as a powerful tool for understanding the complex interactions between pathogens and the human microbiome. This blog explores the utility of microbial metagenomics in the cancer patient population, focusing on infectious disease and molecular diagnostics. Additionally, we will highlight the upcoming MarketsandMarkets Infectious Disease and Molecular Diagnostics Conference, scheduled for October 19th-20th, 2023 in Boston, USA, and discuss the valuable insights attendees can gain from this event.
Understanding Microbial Metagenomics:
Microbial metagenomics is a genomic approach that involves sequencing and analysing the genetic material present in a complex mixture of microorganisms, such as bacteria, viruses, and fungi. This technique provides a comprehensive view of the microbial communities inhabiting a specific environment, such as the human body. By leveraging next-generation sequencing technologies and bioinformatics tools, researchers can identify and characterize the diverse microorganisms present in a sample, even those that are difficult to culture.
Role of Microbial Metagenomics in Infectious Disease Diagnosis:
In cancer patients, diagnosing infectious diseases accurately is crucial due to the potential complications they can cause. Traditional diagnostic methods often rely on culturing specific pathogens, which can be time-consuming and may not always be successful for fastidious or unculturable organisms. Microbial metagenomics offers a promising alternative by allowing for the identification of multiple pathogens simultaneously, without the need for specific prior knowledge or culturing. This approach enables early detection of infections, identification of antibiotic resistance genes, and a deeper understanding of the microbial dynamics within the patient.
Advancements in Molecular Diagnostics:
Microbial metagenomics has paved the way for several innovative molecular diagnostic techniques. These include shotgun metagenomics, which involves sequencing the entire microbial community, and targeted amplicon sequencing, which focuses on specific genomic regions of interest. These methods provide valuable insights into the microbial diversity, pathogenic potential, and antibiotic resistance profiles within a patient sample.
Furthermore, advancements in sequencing technologies and data analysis pipelines have significantly improved the speed and accuracy of microbial metagenomics. Real-time metagenomic sequencing enables rapid identification of pathogens, facilitating prompt treatment decisions and better patient outcomes. Additionally, the integration of metagenomics data with clinical information enhances personalized medicine approaches for cancer patients, ensuring tailored treatment strategies.
MarketsandMarkets Infectious Disease and Molecular Diagnostics Conference 2023:
The forthcoming MarketsandMarkets Infectious Disease and Molecular Diagnostics Conference, scheduled for October 19th-20th, 2023 in Boston, USA, presents a unique opportunity for professionals and researchers in the field. This conference aims to bring together experts from academia, industry, and healthcare to share insights, exchange knowledge, and discuss the latest advancements in the field of infectious disease and molecular diagnostics.
Conference attendees can expect a diverse range of topics to be covered, including:
By attending this conference, participants will gain valuable insights into the latest innovations, network with industry leaders, and foster collaborations with experts in the field. The event promises to foster a vibrant environment for knowledge exchange, driving advancements in infectious disease diagnostics and personalized medicine for cancer patients.
GRAB THE OPPORTUNITY AND REGISTER TODAY!
Takeaways
Microbial metagenomics is revolutionizing infectious disease diagnosis in cancer patients, enabling rapid identification of pathogens and a deeper understanding of the complex interactions between microbes and the human microbiome. The forthcoming MarketsandMarkets Infectious Disease and Molecular Diagnostics Conference provides an excellent platform to explore the latest advancements, exchange ideas, and gain valuable insights from experts in the field. This event will undoubtedly contribute to the continued progress of microbial metagenomics and its translation into clinical practice, ultimately benefiting cancer patients and healthcare providers worldwide.
0 notes
Text
Metagenomics Market Size 2022 Growth Share, Industry Dynamics, Top Trends and Regional Analysis Forecast to 2030
According to a recently released report by Future Market Insights, widespread adoption of metagenomics tools in environmental monitoring and pollution reduction projects is estimated to boost the metagenomics market growth. Recent advancements in technology are aiding in the substantial reduction of costs incurred for sequencing paving way for the development of genetic sequencing on a large scale. This has opened up new opportunities for leveraging metagenomic tools on a large scale for monitoring of environmental microbe communities. Moreover, the efficacy of metagenomic tools in tackling environmental toxicology is further contributing to its growing popularity. For instance, a recent development saw the use of metagenomics in predicting the extent and presence of contamination in the environment while another research outlined how metagenomics can be leveraged to attenuate pollutants in the environment using unculturable microbes present in the natural environment. The use of metagenomics to understand the influence biostimulation can have on microbial communities is also under progress.
Growing concerns about environmental conditions are prompting authorities around the world to invest in researches to improve the efficiency and scale of bioremediation. In addition to this, bioremediation is gradually gaining traction owing to its cost-effective and eco-friendly chemical treatment. As metagenomics is vital to the success of bioremediation, the market is estimated to grow as the alternative chemical treatment process gains popularity.
Clinical Diagnostics to Emerge as a Vital Revenue Pocket
Clinical diagnostics and bioremediation are estimated to emerge as the highest revenue generating segments in the foreseeable future. The term metagenomic diagnostics is slowly gaining in prominence as studies experiment to widen the scope of application of metagenomics in the healthcare industry. For instance, multiple studies have alluded to the development of a new diagnostic procedure termed as “shotgun metagenomics” which uses DNA sequencing to identify different pathogens and aid in the development of effective targeted treatment by predicting their resistance against different medications. Metagenomic diagnostics presents a promising prospect that could help predict and manage the treatment of a plethora of diseases. Further, metagenomics can also potentially discover hidden genetic features that could prove vital in the progression of an assortment of biotechnological applications such as the discovery of novel enzymes, bioactive molecules, and genes with significantly better or altogether new biochemical functionalities.
Metagenomics Market Growth Underpinned by its Potential Application in Antibiotics Production
Metagenomics provides a detailed analysis of the taxonomic composition of microbe communities. It provides accurate information with regards to the gene characteristics specific to a particular community. The information provided by metagenomic tools can be effectively utilized for the production of an assortment of antibiotics. Recent research in the direction was conducted by scientists at UC Berkley who concluded leveraging metagenomics for bioprospecting soil can aid in the development of a plethora of antibiotics. The study used metagenomics to identify the genes that produce antibiotics and antifungals to combat diseases in microorganisms and stated that the same chemicals can potentially aid in combating bacterial and fungal infections in humans as well.
For more information: https://www.futuremarketinsights.com/reports/metagenomics-market
Library Preparation Kits find Growing Consumer Base as Gene Sequencing Gains Centerstage
DNA and RNA sequencing practices are gradually gaining traction owing to their ability to provide a massive set of valuable data about gene sequences in a small duration of time. The data generated through gene sequencing is then analysed by clinicians, scientists, and researchers who then used the derived knowledge in various verticals such as agriculture, forensics, diagnostics, pharmaceuticals, and other applications. Library preparation kits provide the means for efficient and quick extraction of gene segments which are then used for analysis. Moreover, advancements in technology are aiding library kit manufacturers in offering improved products that reduce error and stress while improving accuracy. The FMI report opines the factors will prove vital in the growth of the segment which is estimated to contribute significantly to the metagenomics market growth.
Metagenomics Market Continues to Propel in North America, Upheld by Government Funding for Epigenetics R&D
The FMI study opines metagenomics market in North America is estimated to continue to proliferate and remain at the helm of the global metagenomics market growth. Presence of advanced infrastructure coupled with continuous government funding for research and development in the field of epigenetics are the vital factors that are estimated to drive the growth of the market in the region. Further, the presence of favorable regulations promoting research and development for improving the quality of healthcare in the countries in the region are expected to fuel the market growth.
0 notes
Text
Unveiling the Microbial Enigma: Exploring Modern Techniques for Microbiome Analysis
The microbiome, comprising trillions of microorganisms residing in and on our bodies, holds a vast reservoir of knowledge waiting to be unraveled. Scientists continuously develop innovative methods to study the microbiome, shedding light on its intricate composition and functional dynamics. In this article, we will delve into some of the cutting-edge techniques used to explore and analyze the enigmatic world of the microbiome.
Shotgun Metagenomics
Shotgun metagenomics is a powerful technique that involves sequencing the entire genetic material in a microbial community. This method provides a comprehensive snapshot of microbial diversity, allowing researchers to identify and characterize various microorganisms and their functional genes. By analyzing the collective genetic information, scientists gain insights into the potential roles and interactions of the microbiome in human health and disease.
Metatranscriptomics
Metatranscriptomics focuses on the RNA molecules expressed by microbial communities, providing valuable information about the microbiome's active genes and metabolic activities. By sequencing and analyzing the transcriptome, researchers can determine which genes are actively transcribed and gain insights into the functional dynamics of the microbiome under different conditions or disease states.
Metaproteomics
Metaproteomics involves the large-scale identification and quantification of proteins produced by the microbiome. This technique directly measures the functional proteins within a microbial community, offering insights into the active metabolic pathways and protein-protein interactions. Researchers can link microbial functions to specific biological processes by studying the metaproteome and identifying potential therapeutic targets.
Single-Cell Genomics
Single-cell genomics has emerged as a groundbreaking technique for studying individual microbial cells within a complex community. By isolating and sequencing the DNA of single cells, scientists can capture the genetic information of individual microorganisms, even those that are difficult to culture. This method enables the identification and characterization of rare and unculturable microorganisms, unraveling their unique genetic attributes and functional potential.
Spatial Metagenomics
Spatial metagenomics combines high-resolution imaging techniques with genomic analysis to understand the spatial organization of microbial communities. By visualizing the distribution and interactions of microorganisms within a sample, scientists can identify patterns, spatial dependencies, and potential ecological relationships between different species. This approach provides insights into microorganisms' spatial dynamics and niche specialization within their habitats.
Long-Read Sequencing
Traditional DNA sequencing techniques often produce short DNA fragments, limiting the ability to assemble complete genomes of individual microorganisms. Long-read sequencing technologies, such as Oxford Nanopore and Pacific Biosciences, generate longer DNA sequences, facilitating the reconstruction of complete microbial genomes. This advancement enables a more comprehensive understanding of individual microbial species' genetic potential and functional capabilities.
Integrated Multi-Omics Analysis
To gain a holistic view of the microbiome, scientists are integrating multiple omics approaches, such as genomics, transcriptomics, proteomics, and metabolomics. Researchers can create a comprehensive picture of the microbiome's structure, function, and metabolic activities by combining data from different omics layers. Integrated multi-omics analysis provides a deeper understanding of the intricate interplay between microbial components and their impact on host health.
Conclusion
The microbiome study has undergone remarkable advancements due to the development of innovative techniques. Shotgun metagenomics, metatranscriptomics, metaproteomics, single-cell genomics, spatial metagenomics, long-read sequencing, and integrated multi-omics analysis collectively contribute to unraveling the complexities of the microbial world. These cutting-edge methods enable scientists to explore the functional dynamics, genetic diversity, and ecological relationships within the microbiome, paving the way for novel therapeutic interventions and personalized approaches to healthcare.
0 notes
Text
Paper by former colleagues published on rainbow Trout (Oncorhynchus mykiss) midgut and hindgut microbiomes using whole shotgun metagenomics
As a postdoc at Montana State University in 2015/2016 with Carl Yeoman’s lab, I consulted on a project led by then-PhD student Lola Betiku on a metagenomics dataset from two locations from the digestive tract of trout. Lola is now an Assistant Professor at Florida A&M University, and has been kind enough to continue working on this project to get it published. It was just accepted in the journal…
View On WordPress
1 note
·
View note
Text
Global Microbiome Sequencing Services Market Industry Size, Share, In-Depth Qualitative Insights, Explosive Growth Opportunity, Regional Analysis Forecast
The reports also help in understanding the Global Microbiome Sequencing Services Market Value dynamic, and structure by analyzing the market segments and projecting the Global Microbiome Sequencing Services Market Value. Clear representation of competitive analysis of key players by Design, price, financial position, product portfolio, growth strategies, and regional presence in the Global Microbiome Sequencing Services Market Value makes the report investor's guide.
Global Microbiome Sequencing Services Market Overview:
This Global Microbiome Sequencing Services Marketindustry research provided a comprehensive analysis of the worldwide Global Microbiome Sequencing Services Market, taking into account all critical variables such as growth factors, limitations, market advancements, top investment pockets, future prospects, and trends. The research begins by emphasizing the important trends and possibilities that may develop in the near future and have a favorable influence on overall industry growth.
The Global Microbiome Sequencing Services Market is expected to reach US$ 3.22 Bn. at a CAGR of 5.61% during the forecast period 2029.
Request a Free PDF Sample Report @
Market Scope:
Global Microbiome Sequencing Services Market Research Report analyzed the current state of the definitions, classifications, applications, and industry chain structure. The analysis provides unbiased professional commentary on the present market scenario, prior market performance, production and consumption rates, demand and supply ratios, and income generation forecasts for the projected period. The Global Microbiome Sequencing Services Market study also gives information on the leading businesses functioning in the Global Microbiome Sequencing Services Market industry's strategic ambitions and company growth strategies. Mergers and acquisitions, government and corporate transactions, partnerships and collaborations, joint ventures, brand promotions, and product launches are among the methods evaluated in the research. To summarise what has been said thus far,
The Global Microbiome Sequencing Services Market report presents insights into each of the leading Global Microbiome Sequencing Services Market end users along with annual forecasts to 2027. The report provides revenue forecasts with sales and growth rate of the global Global Microbiome Sequencing Services Market. Forecasts are also provided for the market's product, application, and geographic segments. Forecasts are produced to help people understand the industry's future outlook and potential.
Segmentation:
Based on Technology, Shotgun sequencing segment is expected to witness lucrative growth during the forecast period. Shotgun sequencing's benefits over other technologies, researchers' and healthcare professionals' increasing use of shotgun metagenomic sequencing, and the growing number of metagenomic sequencing-based research activities and investments are all contributing to this rise.
Purchase Inquiry:
Key Players:
The research includes the most recent news and industry developments regarding Global Microbiome Sequencing Services Market expansions, acquisitions, growth strategies, joint ventures and collaborations, product launches, market expansions, and so on. Among the main companies in the Global Microbiome Sequencing Services Market, the sector is
• Baseclear
• Clinical-Microbiomics
• Molzym
• Zymo Research
• Rancho Biosciences
• Microbiome Therapeutics
• Microbiome Insights
• Openbiome
• Resphera Biosciences
• Metabiomics
• Ubiome
• Shanghai Realbio Technology
• Diversigen
• Merieux Nutrisciences
• Second Genome
• Molecular Research LP (MR DNA)
• LOCUS BIOSCIENCES, INC
Regional Analysis:
The primary goal of this study is to assist the user in understanding the market in terms of definition, segmentation, market potential, significant trends, and the problems that the industry is experiencing across ten key regions.
COVID-19 Impact Analysis on Global Microbiome Sequencing Services Market:
The research details the overall impact of COVID-19 on the Health Insurance Market by providing a micro- and macroeconomic analysis. The precise study focuses on market share and size, which clearly depicts the impact that the pandemic has had and is anticipated to have on the global Health Insurance Market in the future years.
Want your Report customized?
Key Questions answered in the Global Microbiome Sequencing Services Market Report are:
What is the function of the Global Microbiome Sequencing Services Market?
What is the predicted revenue generation of the Global Microbiome Sequencing Services Market?
At what growth rate is the Global Microbiome Sequencing Services Market evolving?
Who are the major market giants operating in the Global Microbiome Sequencing Services Market?
About Us:
Maximize Market Research provides B2B and B2C research on 12000 high-growth emerging opportunities & technologies as well as threats to the companies across the Healthcare, Pharmaceuticals, Electronics & Communications, Internet of Things, Food and Beverages, Aerospace and Defense, and other manufacturing sectors .
Contact Us:
MAXIMIZE MARKET RESEARCH PVT. LTD
3rd Floor, Navale IT Park Phase 2,
Pune Banglore Highway,
Narhe, Pune, Maharashtra 411041, India.
Global Microbiome Sequencing Services Market Size, Share, Analysis, Growth, Trends, Drivers, Opportunity
0 notes
Text
Study assesses microbiome of male genital mucosa
Study assesses microbiome of male genital mucosa
There is a strong link between pathophysiology and the human body’s microbiome. Research suggests that many microbial communities regulate health, host homeostasis, disease progression, and infection risk. Recent developments in high throughput sequencing technologies like metagenomic sequencing, the whole genome shotgun, and next-generation sequencing (NGS) techniques have stimulated further…
View On WordPress
0 notes
Text
An Optimized Pipeline for Detection of Salmonella Sequences in Shotgun Metagenomics Datasets
Background: Culture-independent diagnostic tests (CIDTs) are gaining popularity as tools for detecting pathogens in food. Shotgun sequencing holds substantial promise for food testing as it provides abundant information on microbial communities, but the challenge is in analyzing large and complex sequencing datasets with a high degree of both sensitivity and specificity. Falsely classifying sequencing reads as originating from pathogens can lead to unnecessary food recalls or production shutdowns, while low sensitivity resulting in false negatives could lead to preventable illness. Results: We have developed a bioinformatic pipeline for identifying Salmonella as a model pathogen in metagenomic datasets with very high sensitivity and specificity. We tested this pipeline on mock communities of closely related bacteria and with simulated Salmonella reads added to published metagenomic datasets. Salmonella-derived reads could be found at very low abundances (high sensitivity) without false positives (high specificity). Carefully considering software parameters and database choices is essential to avoiding false positive sample calls. With well-chosen parameters plus additional steps to confirm the taxonomic origin of reads, it is possible to detect pathogens with very high specificity and sensitivity. http://dlvr.it/SsyHFt
0 notes