#DNA sequencing
Text
First, let’s address the fact that hackers recently accessed the personal data of about 14,000 23andMe customers. Because of how 23andMe works—it has a “DNA Relatives” feature that lets users find people they are probably related to—this breach created 6.9 million “other users” who had data stolen in the breach, according to reporting by TechCrunch. This data included people’s names, birth year, relationships, percentage of DNA shared with other 23andMe users, and ancestry reports.
[...]
Getting your DNA or your loved ones’ DNA sequenced means you are potentially putting people who are related to those people at risk in ways that are easily predictable, but also in ways we cannot yet predict because these databases are still relatively new. I am writing this article right now because of the hack, but my stance on this issue has been the same for years, for reasons outside of the hack.
In 2016, I moderated a panel at SXSW called “Is Your Biological Data Safe?,” which was broadly about the privacy implications of companies and other entities creating gigantic databases of people’s genetic code. This panel’s experts included a 23andMe executive as well as an FBI field agent. Everyone on the panel and everyone in the industry agrees that genetic information is potentially very sensitive, and the use of DNA to solve crimes is obviously well established.
At the time, many of the possible dangers of providing your genome to a DNA sequencing company were hypothetical. Since then, many of the hypothetical issues we discussed have become a reality in one way or another. For example, on that panel, we discussed the work of an artist who was turning lost strands of hair, wads of chewing gum, and other found DNA into visual genetic “portraits” of people. Last year, the Edmonton Police Service, using a company called Parabon, used a similar process to create 3D images of crime suspects using DNA from the case. The police had no idea if the portrait they generated actually looked like the suspect they wanted, and the practice is incredibly concerning.
To its credit, 23andMe itself has steadfastly resisted law enforcement requests for information, but other large databases of genetic information have been used to solve crimes. Both 23andMe and Ancestry are regularly the recipients of law enforcement requests for data, meaning police do see these companies as potentially valuable data mines.
749 notes
·
View notes
Text
Does anyone else want to do full genomic sequencing on the Saxons and the people of Camelot and then compare them or is it just me?
I want to know how much the two populations diverged in the unspecified time between "Holder of the GRAIL" and Mordred failing his driving test
Also I'm guessing that the evolutionary rate is ridiculously high among Saxons because of the radiations, but also given the low survival rate, in normal conditions there should be a low genetic diversity because of the bottleneck effect
#the mechanisms#the mechs#hnoc#mordred#hnoc mordred#mordred hnoc#saxons hnoc#molecular biology#dna sequencing#i should go to bed#i dont even know if any of this makes sense scientifically
81 notes
·
View notes
Note
CGACTAGCCATCCCTCTGGCTCTTAGATAGCCGGATACAGTGATTTTGAAAGGTTTGTGGGGTACAGCTATGACTTGCTTAGCTGCGTGTGAGGGAAGGAACTTTTGCGTGTTAGTATGTTGACCCGTGTACTACGCATGCGGGTAGATTATGTAGGTTGAGAGATGCAGGAGAAGTTCTCGACCTTCCCGTGGGAGGTGAACCTATTCACTATTGGAGCATTCCGTTCGAGCATGGCAGTAAGTACGCCTTCTCCATTCTGGTAACCTTCATCCCTATCAGAGCTTGGAGCCAATGATCAGGGTTATTCCCTTGGGACAGACTTCCTACTCACAGTCGGTCACATTGGGCTACTCCATGGGTCTTCGGCTTGACCCGGTCTGTTGGGCCGCGATTGCGTGAGTTTCGGCCCCGCGCTGCGCTGTATAGTCGATTCTCATCCGGCCCTCACATCTGGAAACCCCAACTTATTTAGATAACATCATTAGCCGAAGTTGCTGGGCATGTCCACCGTGGAGTCCTCCCCGGGCGTCCCTCCTTCAAATGACGATAAGCACCGGCAAGCACCATTGATCAACGCAAGGATCGGTGATGTTAACAAAGATTCGGCACATTACTCTTGTTGGTGTGGAATCGCTTAACTACGCGGCGAAGCCTTATGGCAAAACCGATGGGGAATGATTCGGGTAGCGCTAAAAGTCCATAGCACGTACATCCCAACCTGGCGTGCGTACAGTTTGACGACCGCTTCACGCTAAGGTGCTGGCCACGTGCTAAATTAATGCGGCTGCACTGCTCTAAGGACAATTACGGAGTGGGCGGCCTGGCGGGAGCACTACCCCATCGACGCGTACTCGAATACTGTATATTGCTCTCACATGAACAAATTAGTAGAGTGCCGCTTTCAGCCCCCCTGTCGTCGGCGACGTCTGTAAAATGGCGTTGATGTGGATCGACTCTATAGAGGCATCTACTGATGCGTAGGGAGATCCGGAATGTA
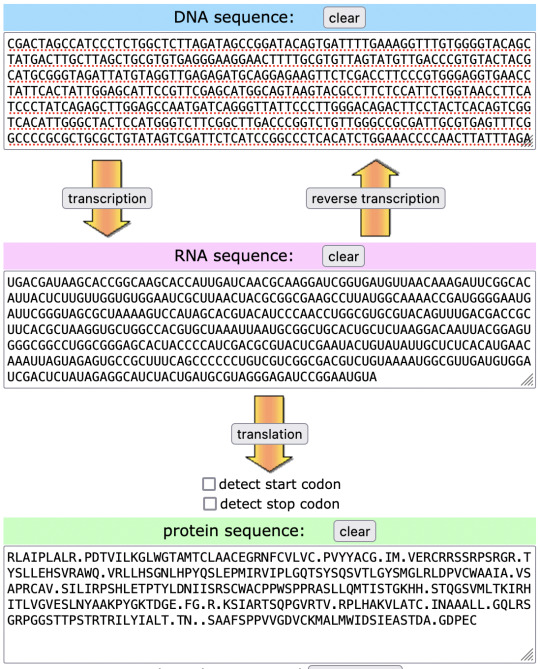
i wish that this was a new ARG cryptography strat for creating messages using only amino acid abbreviations. do better
87 notes
·
View notes
Link
In a monumental milestone, UK Biobank has unveiled open access to incredible whole genome sequencing data for its over half a million participants. This vast genomic resource promises to massively accelerate the diagnosis, treatment, and prevention of diseases worldwide when combined with the study’s expansive health data. Dubbed a “veritable treasure trove” by Professor Sir Rory Collins, Principal Investigator, UK Biobank, the release cements the nonprofit UK Biobank as the most ambitious health research undertaking ever.
Genomic sequencing has revolutionized biomedicine by enabling the reading of the precise DNA code underlying human life. Technologies like PCR unlocked targeted gene testing decades ago. However, the affordable sequencing of entire genomes on a mass scale has only recently become feasible.
While costs have plunged, interpreting genomic data relies heavily on computational analytics. UK Biobank’s dataset amalgamates sequenced genomes from over 350,000 British volunteers with intricate health details like brain imaging, blood assays, lifestyle surveys, and more collected meticulously for 15 years. This unrivaled fusion promises tremendous leaps in illuminating genetics‘ role in disease.
Continue Reading
50 notes
·
View notes
Text








First day for school completed!!! This is all the stuff I made in my classes today >=D
#wander over yonder#woy#save woy#craig mccracken#woy wander#cartoon#my art#my art <3#my artwork#woy wanda#wander over yonder wander#wander over yonder wanda#dna sequencing#biology
25 notes
·
View notes
Text
"With the announcement that the Human Genome Project had mapped all of the genetic material in the human chromosomes, a new era in the understanding of genetics began. The discovery of new genes is announced every day, and it is only a matter of time before the genetic mechanisms of mood disorders is only one of the goals of work in this field. Just as important will be understanding the epigenetic mechanisms by which genes turn on and off and other mechanisms that regulate the expression and work of the instructions encoded in the DNA molecule.
The first genetic approach to pay off in changing treatment is likely to be pharmacogenomics, the field within genetics that investigates genetic factors associated with responses to particular pharmaceuticals rather than with risk of disease. The promise of pharmacogenomics is that therapeutic agents can be rationally selected, based on a person's genetic profile rather than the trial-and-error process patients must now endure. In the not-too-distant future, a blood test will show whether lithium or valproate or lamotrigine or some as yet undiscovered drug will be the best treatment for a particular individual with bipolar disorder. A blood test may be able to identify the bipolar patients who can safely take an antidepressant."
-Francis Mark Mondimore, Bipolar Disorder (2014)
#books#quote#nonfiction#bipolar disorder#mental health#psychology#genomics#dna sequencing#dna#genetics#science#biology
3 notes
·
View notes
Text

DNA is a molecule that carries the genetic information for all living things. It is made of smaller units called nucleotides, which have three parts: a sugar, a phosphate, and a nitrogen base. There are four types of nitrogen bases: adenine (A), thymine (T), cytosine (C), and guanine (G). The nucleotides are linked together in a chain, forming a strand of DNA. Two strands of DNA pair up with each other, forming a double helix. The pairing is based on the rule that A always binds with T and C always binds with G. This way, the two strands are complementary to each other and can store the same information.
DNA is found inside the cells of all organisms. Most of it is located in the nucleus, where it is organized into structures called chromosomes. Each chromosome contains a long piece of DNA that has many genes. Genes are segments of DNA that code for proteins, which are the building blocks and regulators of life. Some DNA is also found in the mitochondria, which are the energy factories of the cell.
DNA is very long compared to its width. A single strand of human DNA is about 5 feet or 1.5 meters long, but it is only 2 nanometers wide. To fit inside the cell, DNA has to be compacted and coiled. It wraps around proteins called histones, forming beads called nucleosomes. The nucleosomes are further coiled into fibers called chromatin, which can condense into chromosomes during cell division.
If we could stretch out all the DNA in one human cell, it would be about 3 km or 6.6 feet long. If we could stretch out all the DNA in all the cells in our body, it would be about twice the diameter of the solar system. That means that our body contains an enormous amount of information encoded in our DNA. This information determines our traits, such as our eye color and height, as well as our risk of diseases and our response to drugs. It also makes us unique from other individuals, except for identical twins who share the same DNA.
DNA is not static; it can change over time due to mutations, which are errors or changes in the sequence of nucleotides. Some mutations are harmless or beneficial, while others are harmful or cause diseases. Mutations can also create variation among individuals and populations, which is the basis of evolution and natural selection.
DNA is a fascinating molecule that reveals a lot about ourselves and our ancestors. By studying DNA, we can learn more about our health, our history, and our future.
3 notes
·
View notes
Text
youtube
Because of an error or a lie, my biological mother assigned me the incorrect ethnicity when she gave me away. It changed the entire course of my life.
Latest painting in my series on what it feels like to be a closed-record adoptee. Watercolor and gouache on hot press paper. Music: Scandinavian Error by Polar Nights, Epidemic Sound
#Watercolor#watercolor painting#watercolour art#gouache#art#female artists#artists on tumblr#making of video#illustration#cuckoo#adoption#adoptee#adoptee voices#closed records#DNA#dna sequencing#heritage#ethnicity#l#Youtube
4 notes
·
View notes
Text


bagong fave sa lab
part ng library prep for DNA sequencing
//
18 notes
·
View notes
Text

There is an entire franchise on why that is a bad idea, 🕛 Anon.

#ask scott lang#scott lang#ant-man#🕛 anon#jurassic park#jurassic world#dinosaurs#dna technology#dna sequencing#cloning#dna cloning#ant man#antman
3 notes
·
View notes
Text
log #11
Are you a base substitution mutation? Because everything you're spewing out right now is nonsense.
4 notes
·
View notes
Text

6 notes
·
View notes
Text
‘Super-spuds’ to the rescue as typical tubers feel the heat

- By Anthony King , Horizon -
From origins in the cool altitudes of the Andes, the potato is not well suited to the extreme temperatures or flooding brought on by climate change. Plant scientists are breeding ‘super-spuds’ able to endure harsher environmental conditions.
The humble potato was first domesticated near Lake Titicaca in present-day Peru at least 8 000 years ago, and went on to sustain the great cities of the Inca empire. By the mid-16th century, it had left the Andes and crossed the Atlantic to Europe where it was introduced to Ireland in 1589 by English adventurer and courtier, the enigmatic Sir Walter Raleigh. Highly productive and extremely popular, the potato plant soon went on to become a staple in many European countries.
Today, it is the fourth most commonly grown food crop globally, after rice, maize, and wheat. Nonetheless, it remains vulnerable to waterlogging and heat stress, conditions that it did not evolve to withstand in its original high-altitude home in the Andes. Now, with pollution upending Europe’s climate, the potato has to confront these dual nemeses with increasing regularity.
‘Some potatoes are quite tolerant of drought stress, but they all have big problems with heat and flooding,’ says Dr Markus Teige, plant scientist at the University of Vienna who is leading the ADAPT project. ADAPT is developing new strategies to ensure potato crop productivity remains stable in the growth conditions of the future.
Plants afflicted by excessive heat stop producing sugars—preventing the development of tubers—and then race to flower early. This is an excellent strategy for wild potatoes to ensure the survival of the species under challenging conditions, but it delivers low yields to farmers.
Climate repercussions
A recent survey of over 500 European potato growers revealed that drought and heat were seen as the main repercussions of climate change on potatoes, followed by pests, disease, and heavy rains.
Some potatoes are quite tolerant of drought stress, but they all have big problems with heat and flooding.
Some potato varieties are better than others at resisting environmental stresses, which suggests that there is potential for plant breeders to genetically improve the European spud to be more tolerant.
The ADAPT project brings together four potato breeders and ten research institutions to investigate how some potatoes resist stresses.
‘We want to understand stress acclimation at the molecular level,’ said Dr Teige, ‘To develop markers for breeding stress tolerant potatoes.’
Potato breeding is especially challenging because of its complex genetics. The European variety contains millions of letters of DNA, each in four copies, on twelve distinct strands (chromosomes).
Genetic markers are akin to signposts that signify important stretches of DNA associated with a desirable trait, such as better tolerance to heat.
‘A relatively small range of potato genetics was brought to Europe,’ said Dr Dan Milbourne, potato researcher at Teagasc in Ireland, a state agricultural research organisation. Therefore, it might be possible to import new traits.
ADAPT scientists have grown around 50 potato varieties in different combinations of stress conditions in various European locations. In parallel, they have run experiments in greenhouses, where varieties are grown under defined conditions in a high-tech facility in the Czech Republic.
It takes about 12 years to produce a potato variety.
The plants are photographed and measured daily to record how much water they use, and their rates of photosynthesis and growth. This data can reveal how they are influenced by stress and highlight signposts (genetic markers) in the potato genome important for stress responses.
The signposts save time and money for future breeding programmes. ‘If a marker is associated with a specific trait, then, when you grow a seedling, you extract the DNA and look for the marker,’ said Dr Teige. The old way was to allow the plant to grow and wait to see if the desired trait was present.
Saving time in plant breeding is a huge deal. ‘It takes about 12 years to produce a potato variety,’ said Dr Milbourne. And he should know, because last year, his organisation was involved in the release of Buster, a new variety of potato resistant to a type of nematode worm that can severely damage potato crops.
Potato preferences
In Ireland, potatoes must be sprayed up to 20 times during a growing season to protect against late blight. Blight has an historical significance in Ireland as it caused potato crop failure in the 1840s which triggered a disastrous famine that decimated the population.
Meanwhile, Europe is seeking to lessen reliance on chemical sprays, with the European Commission recently proposing that pesticide use be cut in half by 2030. To reduce dependence on spraying, more pest-resistant potatoes will be needed.
Dr Milbourne is part of a project called PotatoMASH, which devised a way of scanning the genetic variation across the genome of potato varieties in an inexpensive manner. The method can diagnose the presence of target diseases and pest resistance genes in potatoes by sampling only stretches of very variable DNA, which is significantly less expensive than traditional methods of identifying genetic markers.
New software developed at ILVO (Flanders Research Institute for Agriculture, Fisheries and Food) in Belgium, identifies areas of DNA where there are subtle differences between varieties.
We’re going to have to double production, without increasing the amount of land we farm, while also facing climate change.
It is single differences in the DNA code that are most interesting to breeders, explained Dr Milbourne. Potato breeding will be accelerated by identifying signposts for these areas.
‘Instead of testing thousands of individuals by infecting them with a disease and following their response,’ said Dr Milbourne, ‘I can just click out a small bit of leaf material about the size of my fingernail and test it for these markers, which can tell me whether a gene is present or absent.’
This is an important advance in the push to develop potatoes resistant to pests and diseases and able to withstand the vagaries of our future climate, while not sacrificing yield.
Super-spuds
Crucially, it will not be a matter of breeding just one super-spud, because consumer tastes for potatoes vary widely from country to country, and there will be plenty of new potato varieties needed for the future.
‘We are looking at moving from feeding 7 billion people to between 11 and 13 billion over the next several decades,’ said Dr Milbourne.
‘We’re going to have to double production, without increasing the amount of land we farm, while also facing climate change, which could also deplete the land we have available for agriculture.’
Part of the solution is to boost the resilience of staple crops—such as potatoes—to extremes such as high temperatures, pests, and diseases, while relying less on pesticides. The race is on.
Research in this article was funded by the EU.
This post ‘Super-spuds’ to the rescue as typical tubers feel the heat was originally published on Horizon: the EU Research & Innovation magazine | European Commission.
--
Read Also
Should we genetically edit the food we eat? We asked two experts
#potato#potatoes#plants#genomics#dna#dna sequencing#agriculture#agtech#agritech#climate change#heat#heat wave#drought
1 note
·
View note
Text
Maximizing Efficiency: Best Practices for Using Sequencing Consumables

By implementing these best practices, researchers can streamline sequencing workflows, increase throughput, and achieve more consistent and reproducible results in genetic research. Sequencing Consumables play a crucial role in genetic research, facilitating the preparation, sequencing, and analysis of DNA samples. To achieve optimal results and maximize efficiency in sequencing workflows, it's essential to implement best practices for using these consumables effectively.
Proper planning and organization are essential for maximizing efficiency when using Sequencing Consumables. Before starting a sequencing experiment, take the time to carefully plan out the workflow, including sample preparation, library construction, sequencing runs, and data analysis. Ensure that all necessary consumables, reagents, and equipment are readily available and properly labeled to minimize disruptions and delays during the experiment.
Optimizing sample preparation workflows is critical for maximizing efficiency in sequencing experiments. When working with Sequencing Consumables for sample preparation, follow manufacturer protocols and recommendations closely to ensure consistent and reproducible results. Use high-quality consumables and reagents, and perform regular quality control checks to monitor the performance of the workflow and identify any potential issues early on.
Utilizing automation technologies can significantly increase efficiency when working with Sequencing Consumables. Automated sample preparation systems and liquid handling robots can streamline repetitive tasks, reduce human error, and increase throughput. By automating sample processing and library construction workflows, researchers can save time and resources while improving consistency and reproducibility in sequencing experiments.
Get More Insights On This Topic: Sequencing Consumables
#Sequencing Consumables#DNA Sequencing#Laboratory Supplies#Genetic Analysis#Next-Generation Sequencing#Molecular Biology#Research Tools#Bioinformatics
0 notes
Link
Motivated by the success of the GPT (Generative Pre-trained Transformer) model, researchers from the Southern University of Science and Technology, Tencent AI Lab, Shenzhen, China, and the City University of Hong Kong have developed DNAGPT, a generalized foundation model capable of simultaneously processing multiple DNA sequences from various species. Its unique token design allows users to optimize prompts per their requirements, making it useful in any task. The researchers found that the model improves with pre-training, and it was evaluated on different jobs, including classification (grouping DNA sequences into categories), regression (predicting numerical values), and generation (creating new DNA sequences).
DNA sequences, specifically the large non-coding regions, contain vital, undiscovered information which, when explored, can reveal novel insights underpinning the various mechanisms of life. Different species have similarities along with divergences in their genetic sequences. Therefore, building a generalized tool that can analyze DNA sequences from many different species is imperative.
Continue Reading
42 notes
·
View notes
Text
Introduction to DNA Sequencing and TaqMan Probes
DNA sequencing is a fundamental technique used to determine the order of nucleotides in a DNA molecule, while TaqMan probes are specialized oligonucleotide probes employed in real-time polymerase chain reaction (PCR) assays for the detection and quantification of specific DNA sequences.

Understanding DNA Sequencing
Principles of DNA Sequencing
DNA sequencing relies on various methodologies to decipher the nucleotide sequence of a DNA molecule, providing valuable insights into genetic information and molecular structure.
Importance in Molecular Biology
DNA sequencing plays a crucial role in molecular biology research, enabling scientists to study genetic variation, identify mutations, and unravel the complexities of the genome.
TaqMan Probes: An Overview
Definition and Purpose
TaqMan probes are short, single-stranded DNA probes containing a fluorescent reporter dye and a quencher molecule. These probes hybridize to target DNA sequences during PCR amplification, generating a fluorescent signal proportional to the amount of amplified DNA.
Mechanism of Action
TaqMan probes work by binding to the target DNA sequence between the PCR primers. During PCR amplification, the Taq polymerase cleaves the probe, releasing the reporter dye and generating a fluorescent signal that is detected in real-time.
Applications of DNA Sequencing
Genome Analysis
DNA sequencing is widely used for genome analysis, enabling researchers to sequence entire genomes and study genetic variation, evolutionary relationships, and disease susceptibility.
Mutation Detection
DNA sequencing is invaluable for detecting mutations and genetic alterations associated with diseases such as cancer, inherited disorders, and infectious diseases.
Advantages of TaqMan Probes
Specificity and Sensitivity
TaqMan probes offer high specificity and sensitivity, allowing for the detection of target sequences with unparalleled accuracy and efficiency.
Real-Time PCR Applications
TaqMan probes are compatible with real-time PCR platforms, enabling the quantitative analysis of gene expression, SNP genotyping, and pathogen detection in real-time.
DNA Sequencing Techniques
Sanger Sequencing
Sanger sequencing, also known as chain termination sequencing, is a traditional DNA sequencing method that involves the synthesis of DNA fragments of varying lengths, followed by electrophoretic separation and detection of fluorescently labeled nucleotides.
Next-Generation Sequencing (NGS)
Next-generation sequencing (NGS) technologies enable high-throughput DNA sequencing, allowing for the rapid and cost-effective analysis of entire genomes, transcriptomes, and epigenomes.
Designing and Using TaqMan Probes
Probe Design Considerations
Designing TaqMan probes involves optimizing parameters such as probe length, GC content, and melting temperature to ensure efficient hybridization and specific target detection.
Experimental Applications
TaqMan probes find diverse applications in molecular biology research, including gene expression analysis, SNP genotyping, microbial detection, and viral quantification.
Challenges and Limitations
Cost and Throughput
High cost and limited throughput are significant challenges associated with DNA sequencing and TaqMan probe-based assays, particularly for large-scale genomic studies and diagnostic applications.
Probe Optimization
Optimizing TaqMan probe design and assay conditions is essential for maximizing sensitivity, specificity, and reproducibility, requiring careful experimental optimization and validation.
Conclusion
DNA sequencing and TaqMan probes have revolutionized molecular analysis, enabling researchers to explore the intricacies of the genome and detect specific DNA sequences with unprecedented accuracy and efficiency. By leveraging these technologies, scientists can advance our understanding of genetics and develop innovative solutions for diagnosing and treating genetic diseases.
1 note
·
View note