#gene expression
Text
Ever since the COVID-19 pandemic kicked off, we've been far more aware of our sense of smell.
Now, new research shows that odors – like those emanating from ripening fruits or fermented foods – can lead to changes in how genes are expressed inside cells far beyond the nose.
The findings have scientists wondering if, with much more research, sniffing volatile, airborne compounds could be a way to treat cancer or slow neurodegenerative disease.
Continue Reading.
185 notes
·
View notes
Link
Understanding cell dynamics, regulation, and characteristics has been revolutionized by profiling tests at a previously unheard-of resolution. Nevertheless, the destructive nature of these techniques makes it difficult to monitor the temporal dynamics of living cells. Although it lacks genetic and molecular interpretability, Raman microscopy offers a unique way to report vibrational energy levels at subcellular spatial resolution. The researchers created Raman2RNA (R2R), an experimental and computational framework that uses multi-modal data integration, domain translation, and label-free hyperspectral Raman microscopy images to infer single-cell expression patterns in living cells.
Raman images were used to link scRNA-seq profiles to paired spatial hyperspectral Raman images, and machine learning models were trained to infer expression profiles from Raman spectra at the single-cell level. In reprogramming mouse fibroblasts into induced pluripotent stem cells (iPSCs), R2R accurately inferred the expression patterns of numerous cell stages and fates, including MET cells, iPSCs, stromal cells, fibroblasts, and epithelial cells. This demonstrates how crucial spectroscopic content is to Raman microscopy.
The dynamic balance of extrinsic and internal programs determines the states and functions of cells. Numerous genes work together to coordinate the expression and function of these activities, which include cell proliferation, stress responses, differentiation, and reprogramming. These genes also interact with other cells and the environment to influence these processes.
Continue Reading
44 notes
·
View notes
Text
The neuroscience lecture today was really fun. First about addiction, and then about depression. Discussing the neurochemical changes and how permanent they are. Changes in gene expression and nerve connections. Changes of brain anatomy. Not concerning at all.
....not that I have any of that. Anyway.
#roleplay#roleplayer#unilock roleplay#unilock#sherlock#roleplay blog#college#sherlock holmes#rp#sherlock headcanon#neuroscience#neurochemistry#chemistry major#chemistry#university#biology#gene expression#neuroanatomy#anatomy#science#addiction#depression
13 notes
·
View notes
Photo
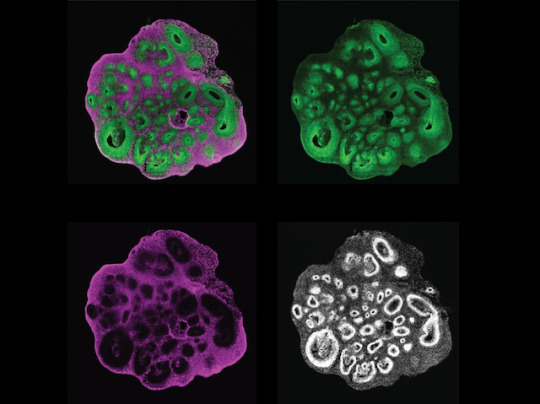
Exploring Family Trees
The cerebral cortex is the outermost layer of your brain, but creating this layered structure is a highly elaborate process during development. It requires early brain cell ancestors – called neural progenitor cells – to differentiate and create later generations of neurons and brain cells called glia. Integral to differentiation is ensuring that genes are switched on at the right time, but also that their genetic instructions are translated into an abundance of proteins to allow progenitors to become brain cells. The importance of these gene-protein networks and their timing during this process is known in mice, but remains unclear in humans. Researchers have now created a human brain organoid (pictured) that allows them to track progenitors (green) and neurons (magenta) simultaneously (all cell nuclei shown in white). This lab-grown ‘mini brain’ allows both cell types from different stages during development to be isolated and analysed, building a more detailed plan of this process.
Written by Sophie Arthur
Image from work by Jaydeep Sidhaye and Philipp Trepte, and colleagues
Institute of Molecular Biotechnology of the Austrian Academy of Sciences (IMBA), Vienna BioCenter (VBC), Vienna, Austria
Image originally published with a Creative Commons Attribution 4.0 International (CC BY 4.0)
Published in eLife, March 2023
You can also follow BPoD on Instagram, Twitter and Facebook
#science#organoids#biomedicine#neuroscience#neural progenitor cells#cells#immunofluorescence#brain#brain cells#neurons#gene expression#embryo development#developmental biology
14 notes
·
View notes
Text
Multiple species of Tardigrades are resistant to drought, high doses of radiation, low oxygen environments, and both high and low temperatures and pressures.
Tardigrades transform into a dormant state called anhydrobiosis, which allows them to reversibly halt their metabolism.
This is a study of Tardigrades genes that allows this.
A large amount of research has focused on the genetic pathways related to these capabilities, and a number of genes have been identified and linked to the extremotolerant response of tardigrades
This study generate the first phylogenies of six separate protein families linked with desiccation and radiation tolerance in Tardigrada
2 notes
·
View notes
Text
New Light on Cells that Break Down Bone - Technology Org
New Post has been published on https://thedigitalinsider.com/new-light-on-cells-that-break-down-bone-technology-org/
New Light on Cells that Break Down Bone - Technology Org
Imaging technology developed at Garvan Institute shows that bone-resorbing osteoclasts gather in distinct pockets, leading to new insights for osteoporosis and cancer treatment.
Image of cells contained in a bone tissue. Image credit: Garvan Institute
Bone may seem like a hard, lifeless structure. Still, the cells living within have been imaged in unprecedented detail, thanks to an innovative imaging method developed at the Garvan Institute of Medical Research.
The new method lets researchers study cells inside the bones of mice, to visualise not just isolated sections, but the entire length of a bone. With a new level of visual detail, the researchers discovered that osteoclasts, cells that break down bone tissue, are more active in some parts of the bone than others.
This knowledge could be used to develop new treatments for osteoporosis, and for dormant cancer cells, which can stay hidden in bone for years until osteoclasts reactivate them.
“Our method has given us an unprecedented window into how cells go about breaking down bone, giving us a new way to investigate osteoporosis and cancer relapse in bone,” says Professor Tri Phan, Head of the Intravital Microscopy Lab and Gene Expression (IMAGE) Lab, immunologist at St Vincent’s Hospital Sydney, Co-Director of the Precision Immunology Program at Garvan and senior author of the paper, published in Nature Protocols.
“We can finally image processes inside bone that we thought were happening, but which were until now beyond the limits of conventional microscopy techniques. We are only beginning to understand the implications of this exciting technology.”
Picture of the bones in a human hand (from an authentic human skeleton). Image credit: Raul654 via Wikimedia, CC-BY-SA-3.0
Giving disease-causing cells no place to hide
Osteoclasts are crucial to the normal maintenance and repair processes of bone, but when they are overly active, they can cause excessive breakdown, known as osteoporosis.
“The inside of living bone is a ‘dark space’ that is difficult to study, because of its hard, mineralised structure,” says co-first author Dr Nayan Deger Bhattacharyya, post-doctoral researcher in the IMAGE Lab. “In order to understand diseases such as osteoporosis and cancer recurrence, we’ve needed to develop the technology to look inside bone tissue.”
The new technique developed at Garvan’s ACRF INCITe Centre can image other dynamic cellular processes until now hidden in bone.
“Our new imaging method is minimally invasive and lets us map out localised populations of cells along the length of an entire bone in our mouse models, instead of just in small sections,” says co-first author Wunna Kyaw, PhD student in the IMAGE Lab.
The researchers tracked down distinct pockets of bone resorption activity as the cells ‘morph’ between actively resorbing osteoclasts and an intermediate cell state called osteomorphs, in real time.
Osteoporosis in bones. Image credit: Scientific Animations, CC BY-SA 4.0
“We suspect these osteomorphs are dangerous as they can accumulate while osteoporosis treatment is administered but can rapidly reform activated osteoclasts to supercharge bone breakdown as soon as treatment is stopped.”
“This would explain an observation in the clinic, that many osteoporosis patients taking the medication denosumab, which blocks osteoclasts from resorbing bone, experience rebound vertebral fractures after they stop using the drug. We will use our imaging method to study how this withdrawal effect could be prevented,” says co-author Professor Peter Croucher, Head of the Bone Biology Lab at Garvan.
The researchers say their method could also be used to investigate cancer cells that can migrate to bone during cancer treatment and lie dormant there for years, only to be reactivated by osteoclasts breaking down the bone tissue surrounding them.
“Being able to see cells and molecules interact in the bone – and one day target them – could be a critical new tool for bone-related diseases,” says Professor Phan.
Source: Garvan Institute
You can offer your link to a page which is relevant to the topic of this post.
#Aging news#animations#Biology#Biotechnology news#Cancer#cancer cells#cancer treatment#cell#cell biology#Cells#Dark#Disease#Diseases#drug#gene expression#Giving#hand#Health & medicine news#how#human#Imaging#immunology#insights#Light#Link#map#Medical devices#medical research#Method#mice
2 notes
·
View notes
Text
As fetuses, we are all phenotypically female until about the 6th or 7th week of gestation, when, in some cases, changes in gene expression on the Y chromosome result in the development of testes.

#facts#interesting facts#humans#human pregnancy#pregnancy#pregnancy facts#gender#y chromosome#gene expression#science#biology#human fetus
7 notes
·
View notes
Text
Born Again🐐🧬
By:
0 notes
Text
FISH Hybridization System
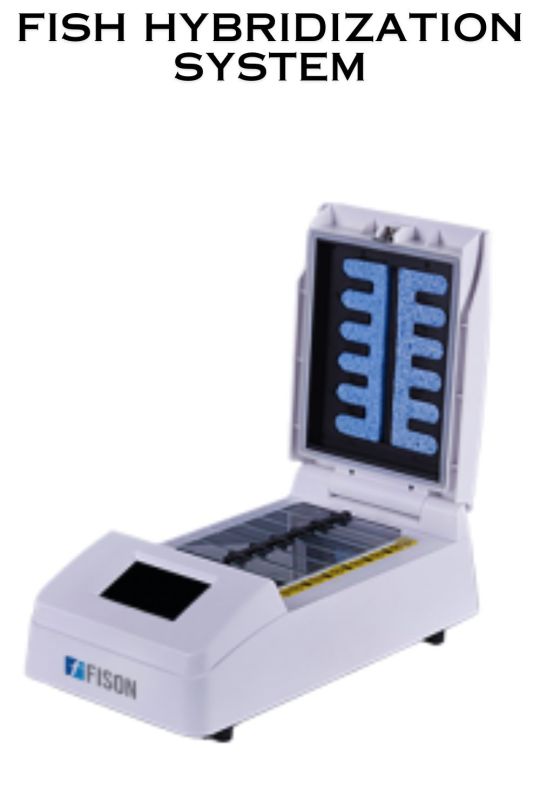
A FISH (Fluorescence In Situ Hybridization) Hybridization System is a specialized laboratory instrument used for detecting and analyzing specific DNA or RNA sequences within cells or tissues. Highly-efficient, stable and convenient operation.
0 notes
Text
Unlocking the Mysteries of Gene Expression: From Genomic Imprinting to Non-Coding RNAs in Biology Class with Dr. Mishra
Unlocking the Mysteries of Gene Expression: From Genomic Imprinting to Non-Coding RNAs in Biology Class with Dr. Mishra #GeneExpression #BiologyClass #GenomicImprinting #NonCodingRNAs #CollaborativeLearning
Dr. Mishra: “Hello there! I see we have a new face in our biology class today. Welcome! We’re so glad you’re here, and I want you to know that this classroom is a friendly and supportive place. If you ever have questions or need assistance, don’t hesitate to ask me or your classmates. We’re all here to learn together and make this an enjoyable experience for you.”
Abby: “Hello. My name is Abha…

View On WordPress
#biology#Chromatin Structure#Collaborative Learning#Epigenetic Modifications#Gene expression#Genomic Imprinting#MicroRNAs#Non-Coding RNAs#Transcription Factors
0 notes
Text
Drug resistance in multiple myeloma: When cancer cells say "NO" to treatment
Drug resistance is like a game of cat and mouse. Cancer cells are the cat, and researchers are the mouse. The cat is always trying to find new ways to catch the mouse, but the mouse is always trying to find new ways to avoid getting caught.
Read More
#multiple myeloma#molecular mechanisms#signaling pathways#Health#stem cell transplantation#oncogenes#Lifestyle#cancer cells#gene mutations#drug resistance#cellular environment#gene expression
0 notes
Link
The dynamic localization of thousands of unique regulatory proteins to specific DNA sequences regulates the expression of genes. It has been a longstanding objective of molecular biology to comprehend this cell-type specific mechanism. However, it is still difficult because the majority of DNA-protein mapping techniques only look at one protein at a time.
A study was conducted to overcome this problem by a group of researchers who introduced a split-pool-based technique called ChIP-DIP (ChIP Done In Parallel), which allows hundreds of different regulatory proteins to be simultaneously mapped throughout the genome in a single experiment. Researchers show that all classes of DNA-associated proteins, such as transcription factors, RNA polymerases, chromatin regulators, and histone modifications, produce extremely accurate maps when generated by ChIP-DIP. By analyzing these data, scientists can identify different types of regulatory elements and their functional activity by defining quantitative combinations of protein localization on genomic DNA.
Thousands of regulatory proteins that localize at specific DNA sequences to activate, repress, and modulate transcription levels are involved in the regulation of gene expression specific to cell types. The Cell-type-specific chromatin states are defined by histone modifications and chromatin states, which are managed by chromatin regulators. These regulators regulate nucleosome location, DNA accessibility, and the reading, writing, and erasing of particular histone modifications. Molecular biology has spent decades trying to figure out how regulatory protein binding results in the expression of genes that are particular to a given cell type. The cell-type specificity of regulatory proteins and histone modifications’ interactions makes genome-wide mapping of these alterations difficult.
Continue Reading
44 notes
·
View notes
Text

#anti ageing treatment#brandidentity#logo design#brand design#Copyright#design#Biotechnology#pharmaceutical#epigenetics#gene expression#science
1 note
·
View note
Photo

Sight Mapping
Identifying the tiny changes in DNA that might be causing disease is like trying to spot a single typo in a whole book. Figuring out how that change is having an impact is even harder. Researchers taking up this challenge for adult macular degeneration – a leading cause of blindness – focussed on cells in the eye called the retinal pigmented epithelium (RPE). They explored a protein called LHX2 (red in the composite image of mouse eye structures and lab-grown human cells) that influences gene expression in the RPE. Without LHX2, protein production dropped overall, and by mapping where LHX2 and another related protein (green) bind to DNA, they managed to pinpoint key genes involved in adult macular degeneration. Variation in the DNA influences how well LHX2 can bind, altering important gene expression and revealing details of macular degeneration risk, and potentially steering future research into new treatment approaches.
Written by Anthony Lewis
Images by Mazal Cohen-Gulkar, composite by Ruth Ashery-Padan
Research by Mazal Cohen-Gulkar, Ahuvit David, Naama Messika-Gold et al, Department of Human Molecular Genetics and Biochemistry, Sackler Faculty of Medicine and Sagol School of Neurosciences, Tel Aviv University, Tel Aviv, Israel
Image originally published with a Creative Commons Attribution 4.0 International (CC BY 4.0)
Research published in PLOS Biology, January 2023
You can also follow BPoD on Instagram, Twitter and Facebook
#science#biomedicine#neuroscience#retina#gene expression#age-related macular degeneration#macular degeneration#blindness#eyesight#immunofluorescence
17 notes
·
View notes
Text
Not long ago, scientists referred to noncoding stretches of DNA as "junk DNA". However, scientists have learned that noncoding stretches of DNA play a critical role in controlling the expression of other genes, often while residing at large distance from the genes, and that mutating, silencing or rewiring these enhancers can cause disease.
By mapping 3D interactions, we better understanding of what controls gene expression and how genes can coordinately change their levels during the transition between different cell fates.
DNA is what controls Gene expression in cells, stored in cell nucleus. DNA and environmental factors determine how your body works.
Non-coding regulatory elements through which transcriptional regulators enact these fates remain understudied.
At a genome-wide scale, enhancer activity and 3D connectivity in embryo-derived stem cell lines that represent each of the early developmental fates.
We observe extensive enhancer remodeling and fine-scale 3D chromatin rewiring among the three lineages, which strongly associate with transcriptional changes, a
#molecular biology#dna#genetics#chromatin#gene expression#epigenetics#cells#science article#scientific research
1 note
·
View note
Text
Circadian rhythms can influence drugs’ effectiveness
New Post has been published on https://thedigitalinsider.com/circadian-rhythms-can-influence-drugs-effectiveness/
Circadian rhythms can influence drugs’ effectiveness


Giving drugs at different times of day could significantly affect how they are metabolized in the liver, according to a new study from MIT.
Using tiny, engineered livers derived from cells from human donors, the researchers found that many genes involved in drug metabolism are under circadian control. These circadian variations affect how much of a drug is available and how effectively the body can break it down. For example, they found that enzymes that break down Tylenol and other drugs are more abundant at certain times of day.
Overall, the researchers identified more than 300 liver genes that follow a circadian clock, including many involved in drug metabolism, as well as other functions such as inflammation. Analyzing these rhythms could help researchers develop better dosing schedules for existing drugs.
“One of the earliest applications for this method could be fine-tuning drug regimens of already approved drugs to maximize their efficacy and minimize their toxicity,” says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT, and a member of MIT’s Koch Institute for Integrative Cancer Research and the Institute for Medical Engineering and Science (IMES).
The study also revealed that the liver is more susceptible to infections such as malaria at certain points in the circadian cycle, when fewer inflammatory proteins are being produced.
Bhatia is the senior author of the new study, which appears today in Science Advances. The paper’s lead author is Sandra March, a research scientist in IMES.
Metabolic cycles
It is estimated that about 50 percent of human genes follow a circadian cycle, and many of these genes are active in the liver. However, exploring how circadian cycles affect liver function has been difficult because many of these genes are not identical in mice and humans, so mouse models can’t be used to study them.
Bhatia’s lab has previously developed a way to grow miniaturized livers using liver cells called hepatocytes, from human donors. In this study, she and her colleagues set out to investigate whether these engineered livers have their own circadian clocks.
Working with Charles Rice’s group at Rockefeller University, they identified culture conditions that support the circadian expression of a clock gene called Bmal1. This gene, which regulates the cyclic expression of a wide range of genes, allowed the liver cells to develop synchronized circadian oscillations. Then, the researchers measured gene expression in these cells every three hours for 48 hours, enabling them to identify more than 300 genes that were expressed in waves.
Most of these genes clustered in two groups — about 70 percent of the genes peaked together, while the remaining 30 percent were at their lowest point when the others peaked. These included genes involved in a variety of functions, including drug metabolism, glucose and lipid metabolism, and several immune processes.
Once the engineered livers established these circadian cycles, the researchers could use them to explore how circadian cycles affect liver function. First, they set out to study how time of day would affect drug metabolism, looking at two different drugs — acetaminophen (Tylenol) and atorvastatin, a drug used to treat high cholesterol.
When Tylenol is broken down in the liver, a small fraction of the drug is converted into a toxic byproduct known as NAPQI. The researchers found that the amount of NAPQI produced can vary by up to 50 percent, depending on what time of day the drug is administered. They also found that atorvastatin generates higher toxicity at certain times of day.
Both of these drugs are metabolized in part by an enzyme called CYP3A4, which has a circadian cycle. CYP3A4 is involved in processing about 50 percent of all drugs, so the researchers now plan to test more of those drugs using their liver models.
“In this set of drugs, it will be helpful to identify the time of the day to administer the drug to reach the highest effectiveness of the drug and minimize the adverse effects,” March says.
The MIT researchers are now working with collaborators to analyze a cancer drug they suspect may be affected by circadian cycles, and they hope to investigate whether this may also be true of drugs used in pain management.
Susceptibility to infection
Many of the liver genes that show circadian behavior are involved in immune responses such as inflammation, so the researchers wondered if this variation might influence susceptibility to infection. To answer that question, they exposed the engineered livers to Plasmodium falciparum, a parasite that causes malaria, at different points in the circadian cycle.
These studies revealed that the livers were more likely to become infected after exposure at different times of day. This is due to variations in the expression of genes called interferon-stimulated genes, which help to suppress infections.
“The inflammatory signals are much stronger at certain times of days than others,” Bhatia says. “This means that a virus like hepatitis or parasite like the one that causes malaria might be better at taking hold in your liver at certain times of the day.”
The researchers believe this cyclical variation may occur because the liver dampens its response to pathogens following meals, when it is typically exposed to an influx of microorganisms that might trigger inflammation even if they are not actually harmful.
Bhatia’s lab is now taking advantage of these cycles to study infections that are usually difficult to establish in engineered livers, including malaria infections caused by parasites other than Plasmodium falciparum.
“This is quite important for the field, because just by setting up the system and choosing the right time of infection, we can increase the infection rate of our culture by 25 percent, enabling drug screens that were otherwise impractical,” March says.
The research was funded by the MIT International Science and Technology Initiatives MIT-France program, the Koch Institute Support (core) Grant from the U.S. National Cancer Institute, the National Institute of Health and Medical Research of France, and the French National Research Agency.
#applications#Behavior#Cancer#Cells#circadian clock#circadian cycle#computer#Computer Science#drug#drugs#effects#Electrical Engineering&Computer Science (eecs)#engineering#enzyme#enzymes#Fraction#France#gene expression#genes#Giving#glucose#Health#Health sciences and technology#hepatitis#how#human#humans#infection#infections#inflammation
0 notes